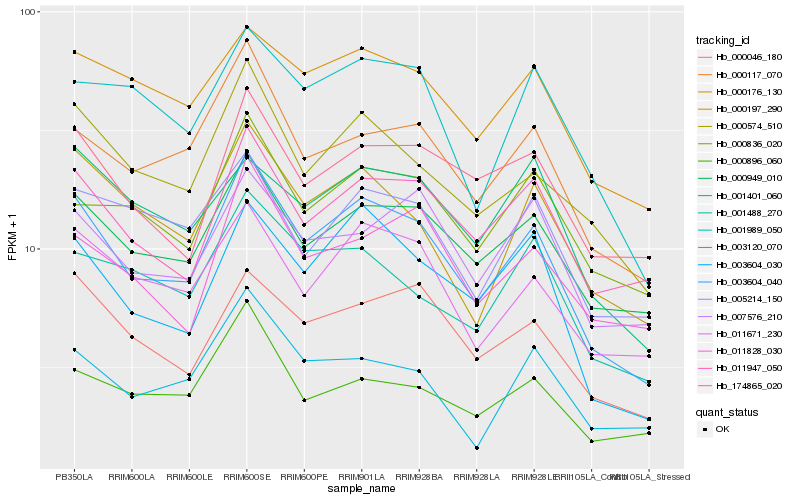
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003604_040 |
0.0 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 2 |
Hb_000574_510 |
0.084892427 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000949_010 |
0.0863713111 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_003604_030 |
0.0873418659 |
- |
- |
hypothetical protein MIMGU_mgv1a025764mg, partial [Erythranthe guttata] |
| 5 |
Hb_000117_070 |
0.0917849072 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 6 |
Hb_011671_230 |
0.0924169142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000046_180 |
0.0948596918 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 8 |
Hb_000197_290 |
0.0952706826 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 9 |
Hb_011947_050 |
0.0955445387 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 10 |
Hb_000176_130 |
0.1023601747 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 11 |
Hb_174865_020 |
0.106489129 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 12 |
Hb_001989_050 |
0.1071672676 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 13 |
Hb_001488_270 |
0.1077719745 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 14 |
Hb_000836_020 |
0.1083173161 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 15 |
Hb_003120_070 |
0.1107067641 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000896_060 |
0.1114534953 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_007576_210 |
0.1115881679 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 18 |
Hb_005214_150 |
0.1116550737 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 19 |
Hb_001401_060 |
0.1132606616 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 20 |
Hb_011828_030 |
0.1147549015 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |