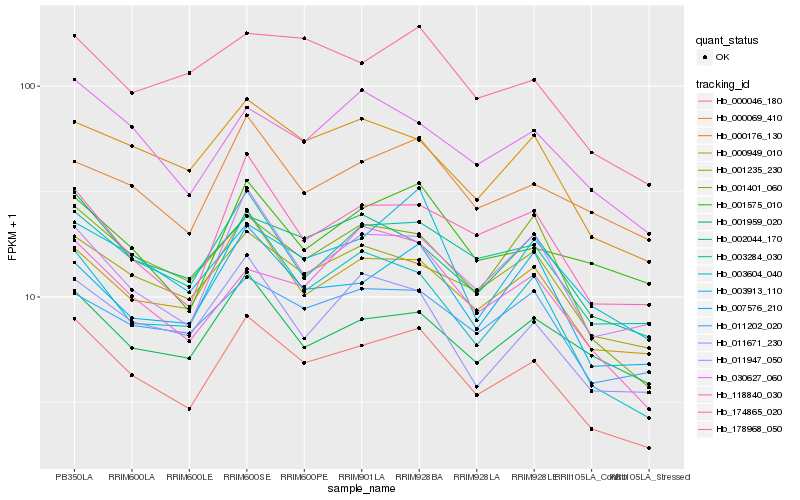
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 2 |
Hb_003284_030 |
0.080576033 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000949_010 |
0.0825138284 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_001401_060 |
0.0880256994 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 5 |
Hb_174865_020 |
0.0887052286 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 6 |
Hb_011947_050 |
0.0902637253 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 7 |
Hb_002044_170 |
0.0902735173 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 8 |
Hb_007576_210 |
0.0903963325 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 9 |
Hb_000176_130 |
0.0911378927 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 10 |
Hb_001235_230 |
0.0942102997 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 11 |
Hb_003604_040 |
0.0948596918 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 12 |
Hb_011671_230 |
0.0964270512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_030627_060 |
0.0975729561 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 14 |
Hb_003913_110 |
0.0986483227 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 15 |
Hb_011202_020 |
0.1020940824 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_118840_030 |
0.1021779443 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 17 |
Hb_001959_020 |
0.1026371245 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 18 |
Hb_178968_050 |
0.1034952755 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 19 |
Hb_001575_010 |
0.103743573 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000069_410 |
0.1055395991 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |