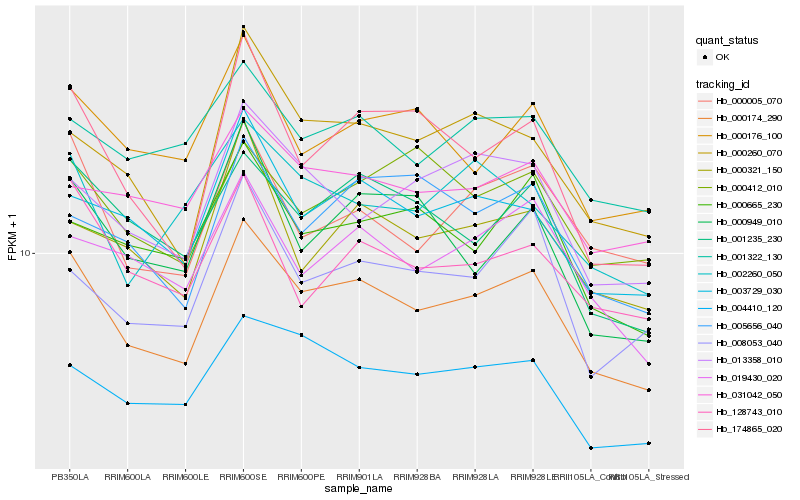
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_290 |
0.0 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_003729_030 |
0.0718340874 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 3 |
Hb_001235_230 |
0.0731047145 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 4 |
Hb_004410_120 |
0.0741845797 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000005_070 |
0.0771973625 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_019430_020 |
0.078896677 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001322_130 |
0.0798160059 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 8 |
Hb_013358_010 |
0.079988719 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 9 |
Hb_174865_020 |
0.0801024683 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 10 |
Hb_000176_100 |
0.0802382178 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 11 |
Hb_000260_070 |
0.0809461006 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000321_150 |
0.0830713748 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000665_230 |
0.0835191495 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 14 |
Hb_008053_040 |
0.083923776 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 15 |
Hb_128743_010 |
0.0846902758 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000412_010 |
0.0854533878 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_005656_040 |
0.0861365148 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_000949_010 |
0.0863681691 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 19 |
Hb_031042_050 |
0.0864526613 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 20 |
Hb_002260_050 |
0.0879617107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |