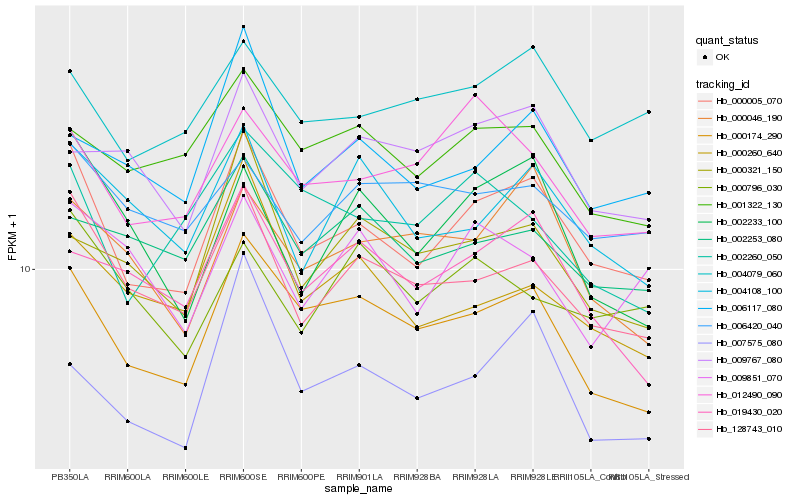
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_070 |
0.0 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_128743_010 |
0.0725312824 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000174_290 |
0.0771973625 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_004079_060 |
0.0950127014 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 5 |
Hb_004108_100 |
0.0966510292 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 6 |
Hb_006117_080 |
0.098490601 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002233_100 |
0.099419113 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 8 |
Hb_012490_090 |
0.1011072228 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 9 |
Hb_000046_190 |
0.101483774 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 10 |
Hb_001322_130 |
0.1027236463 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 11 |
Hb_019430_020 |
0.1034297439 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000260_640 |
0.104914175 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 13 |
Hb_000321_150 |
0.1063384723 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007575_080 |
0.1063672286 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 15 |
Hb_006420_040 |
0.1079360908 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002253_080 |
0.108587889 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 17 |
Hb_009767_080 |
0.1086871164 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 18 |
Hb_009851_070 |
0.1092142517 |
- |
- |
PREDICTED: cyclin-dependent kinase E-1 [Jatropha curcas] |
| 19 |
Hb_000796_030 |
0.1093943786 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_002260_050 |
0.1098833644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |