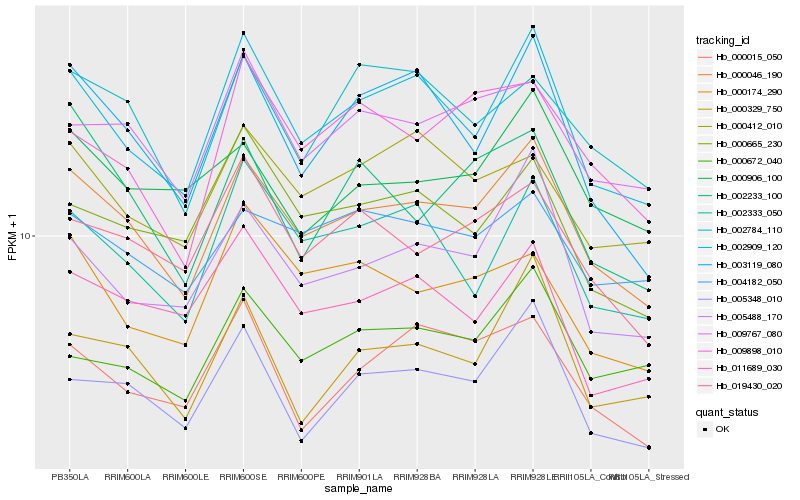
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 2 |
Hb_002909_120 |
0.0558659776 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 3 |
Hb_009767_080 |
0.0757803299 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 4 |
Hb_003119_080 |
0.0760337988 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 5 |
Hb_019430_020 |
0.0761223071 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005488_170 |
0.0766388119 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_005348_010 |
0.0770439138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004182_050 |
0.0793572878 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002333_050 |
0.080783427 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002784_110 |
0.0830074909 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 11 |
Hb_000906_100 |
0.08451006 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 12 |
Hb_000412_010 |
0.0845454997 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000665_230 |
0.0863855381 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 14 |
Hb_000329_750 |
0.0866569992 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 15 |
Hb_011689_030 |
0.0870446637 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 16 |
Hb_000672_040 |
0.0879173484 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 17 |
Hb_000174_290 |
0.0883575094 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_000015_050 |
0.0886310175 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 19 |
Hb_009898_010 |
0.0886316128 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 20 |
Hb_002233_100 |
0.0891201236 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |