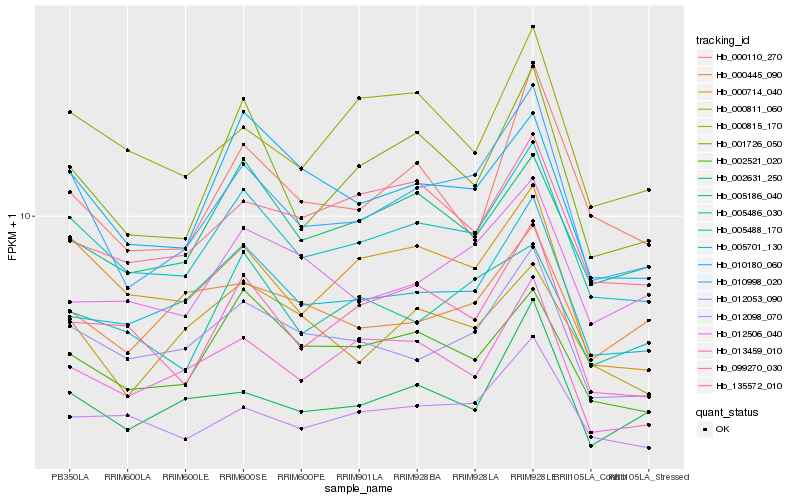
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010998_020 |
0.0 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 2 |
Hb_005488_170 |
0.0705312042 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 3 |
Hb_012053_090 |
0.0977005129 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 4 |
Hb_001726_050 |
0.0991971391 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_005701_130 |
0.1035624306 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 6 |
Hb_000714_040 |
0.1043505148 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002631_250 |
0.106689454 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 8 |
Hb_012098_070 |
0.106955512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 9 |
Hb_000110_270 |
0.1139284955 |
- |
- |
prp4, putative [Ricinus communis] |
| 10 |
Hb_010180_060 |
0.1166112568 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 11 |
Hb_000811_060 |
0.1174417283 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 12 |
Hb_013459_010 |
0.1174555412 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 13 |
Hb_012506_040 |
0.1197523926 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 14 |
Hb_099270_030 |
0.120103236 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000815_170 |
0.1222897161 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 16 |
Hb_000445_090 |
0.1236335189 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 17 |
Hb_002521_020 |
0.1237963491 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_005486_030 |
0.1254205129 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 19 |
Hb_005186_040 |
0.1257486935 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 20 |
Hb_135572_010 |
0.1258203614 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |