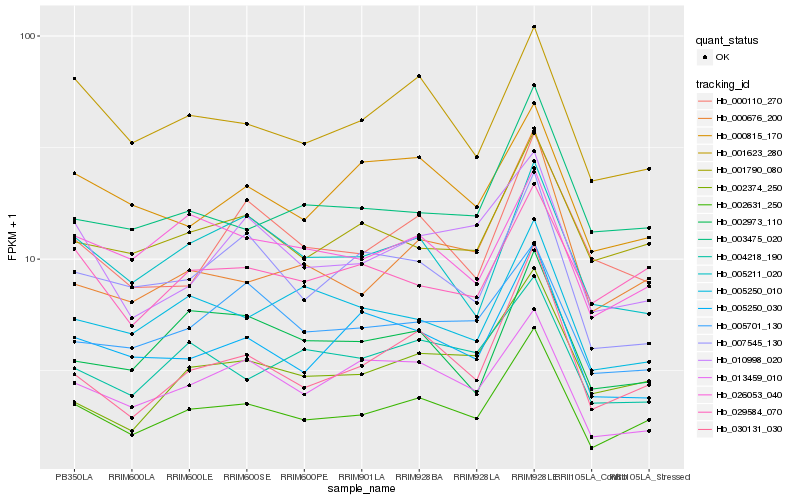
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_250 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 2 |
Hb_030131_030 |
0.080948878 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000676_200 |
0.094981632 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 4 |
Hb_001790_080 |
0.0972339131 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_003475_020 |
0.0982923464 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 6 |
Hb_029584_070 |
0.1027491002 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 7 |
Hb_001623_280 |
0.1030011074 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005250_030 |
0.1040469581 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 9 |
Hb_010998_020 |
0.106689454 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 10 |
Hb_004218_190 |
0.1070669587 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007545_130 |
0.1070849326 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002374_250 |
0.1078824907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_013459_010 |
0.110382381 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 14 |
Hb_005211_020 |
0.1104040531 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000110_270 |
0.1117553437 |
- |
- |
prp4, putative [Ricinus communis] |
| 16 |
Hb_026053_040 |
0.1122730476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005250_010 |
0.112761341 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 18 |
Hb_005701_130 |
0.1132307912 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 19 |
Hb_002973_110 |
0.1142850586 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000815_170 |
0.115383054 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |