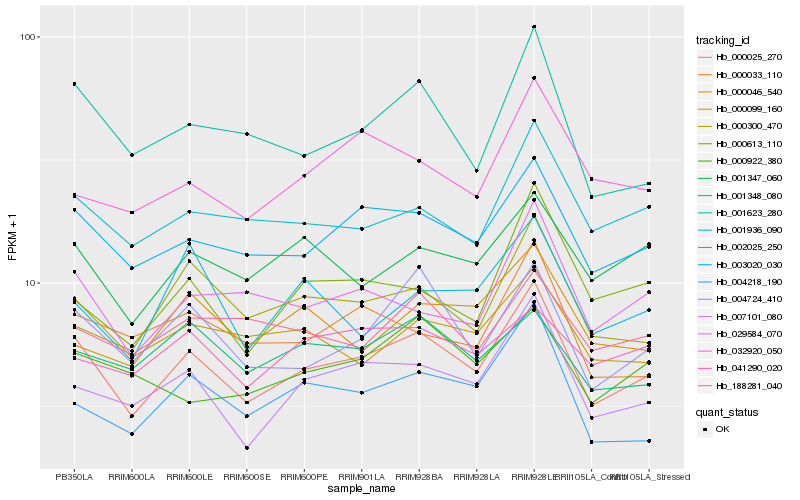
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_270 |
0.0 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_003020_030 |
0.0711018661 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_007101_080 |
0.0824247176 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 4 |
Hb_004724_410 |
0.0884909707 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001623_280 |
0.0905051262 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001936_090 |
0.0947460409 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004218_190 |
0.0970814951 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001347_060 |
0.0999009337 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 9 |
Hb_029584_070 |
0.1001816745 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 10 |
Hb_000099_160 |
0.1012070566 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 11 |
Hb_000613_110 |
0.1033349079 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 12 |
Hb_000300_470 |
0.1035510683 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 13 |
Hb_000033_110 |
0.103931688 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 14 |
Hb_032920_050 |
0.1093589058 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001348_080 |
0.1102298838 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 16 |
Hb_188281_040 |
0.1125929131 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 17 |
Hb_000922_380 |
0.112685672 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein MPP10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_540 |
0.1129380801 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 19 |
Hb_002025_250 |
0.113205214 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 20 |
Hb_041290_020 |
0.113301212 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |