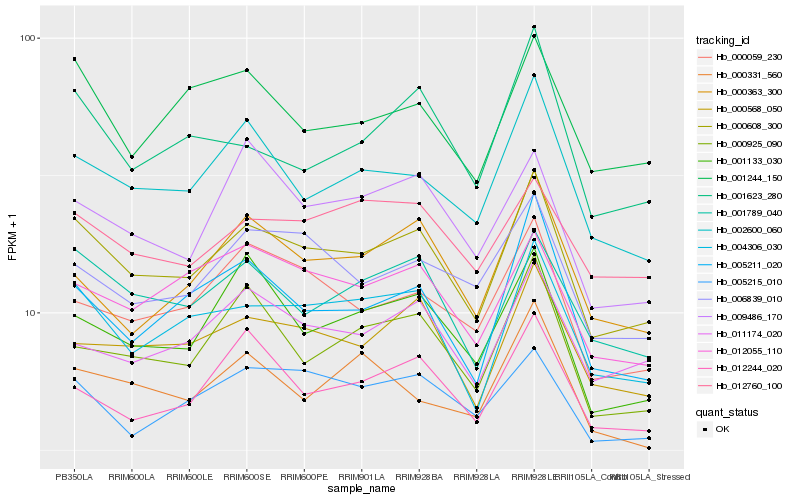
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_300 |
0.0 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 2 |
Hb_004306_030 |
0.056691178 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 3 |
Hb_001789_040 |
0.063691817 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000568_050 |
0.0691054539 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002600_060 |
0.0707505435 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 6 |
Hb_005211_020 |
0.0749517768 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_012055_110 |
0.0762505411 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 8 |
Hb_000925_090 |
0.0768319934 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001133_030 |
0.0779146625 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 10 |
Hb_001244_150 |
0.0785068418 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 11 |
Hb_000059_230 |
0.0788837906 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 12 |
Hb_006839_010 |
0.0792547736 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 13 |
Hb_011174_020 |
0.0796892465 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 14 |
Hb_012244_020 |
0.0802310501 |
- |
- |
calpain, putative [Ricinus communis] |
| 15 |
Hb_012760_100 |
0.082350047 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 16 |
Hb_005215_010 |
0.0828267692 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001623_280 |
0.0846023029 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_009486_170 |
0.0862137421 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 19 |
Hb_000363_300 |
0.0873412036 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000331_560 |
0.087391114 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |