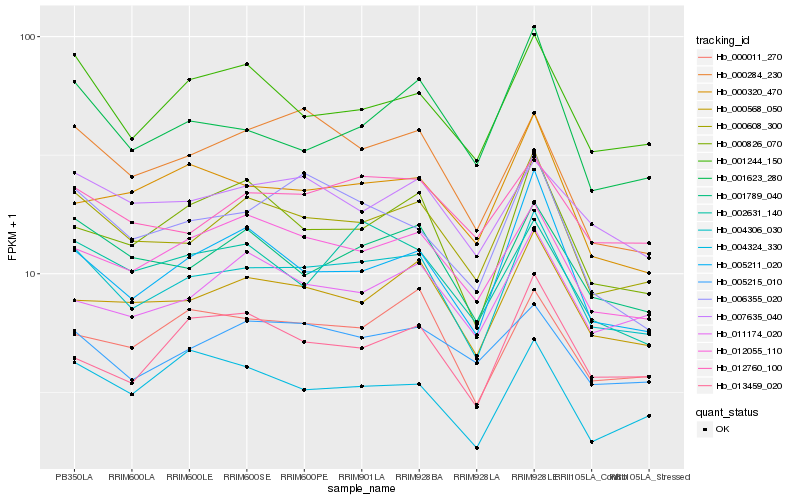
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_030 |
0.0 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 2 |
Hb_000608_300 |
0.056691178 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 3 |
Hb_001789_040 |
0.0724213599 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000568_050 |
0.0747419178 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001244_150 |
0.0808394423 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 6 |
Hb_012055_110 |
0.0817991508 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 7 |
Hb_000011_270 |
0.0829078815 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 8 |
Hb_005211_020 |
0.0837599919 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000826_070 |
0.0853142981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012760_100 |
0.0862081731 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 11 |
Hb_007635_040 |
0.0868874767 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006355_020 |
0.0883050835 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 13 |
Hb_000284_230 |
0.0883553167 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 14 |
Hb_002631_140 |
0.0894108318 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005215_010 |
0.090121983 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001623_280 |
0.0919622205 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000320_470 |
0.0930210194 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 18 |
Hb_011174_020 |
0.0935803585 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 19 |
Hb_004324_330 |
0.0945084797 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 20 |
Hb_013459_020 |
0.0956347613 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |