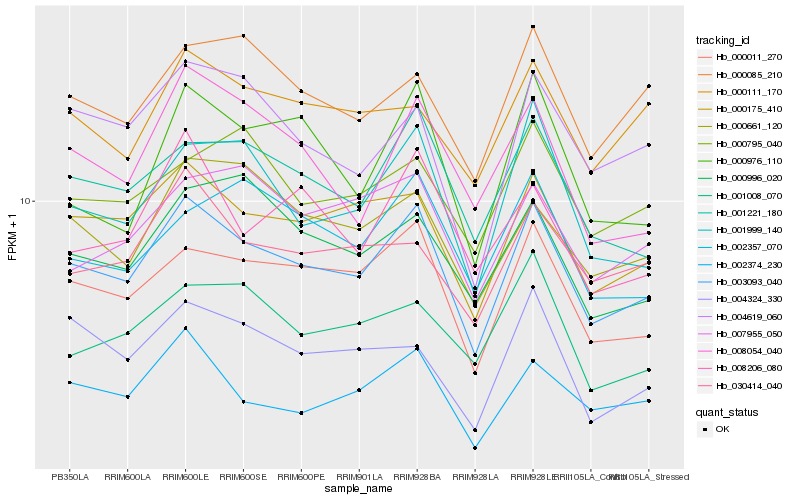
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003093_040 |
0.0 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000011_270 |
0.0569261589 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 3 |
Hb_000175_410 |
0.062601556 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_001221_180 |
0.0694246981 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_001999_140 |
0.0749513378 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_002374_230 |
0.0750769356 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 7 |
Hb_000661_120 |
0.0816857129 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 8 |
Hb_004619_060 |
0.0838595114 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 9 |
Hb_008054_040 |
0.0839228272 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 10 |
Hb_000085_210 |
0.0851333935 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 11 |
Hb_030414_040 |
0.0859288794 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000996_020 |
0.0861456161 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 13 |
Hb_002357_070 |
0.0889958395 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 14 |
Hb_000976_110 |
0.0895933874 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 15 |
Hb_000111_170 |
0.0896331535 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000795_040 |
0.0898001208 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_007955_050 |
0.0898267428 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_004324_330 |
0.090224227 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 19 |
Hb_008206_080 |
0.0905575871 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 20 |
Hb_001008_070 |
0.0905988908 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |