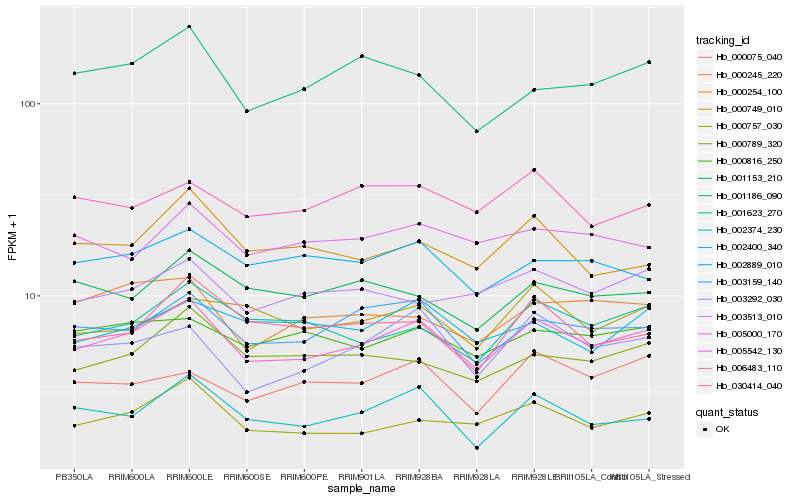
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 2 |
Hb_003159_140 |
0.066152104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000757_030 |
0.0725303175 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 4 |
Hb_002374_230 |
0.0786239488 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 5 |
Hb_003292_030 |
0.0800522718 |
- |
- |
PREDICTED: exosome complex component RRP42 [Populus euphratica] |
| 6 |
Hb_001623_270 |
0.0807317243 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 7 |
Hb_000789_320 |
0.084125943 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000816_250 |
0.0846307897 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002889_010 |
0.0866722489 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 10 |
Hb_030414_040 |
0.0868143476 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003513_010 |
0.0871456029 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 12 |
Hb_001186_090 |
0.0884833072 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 13 |
Hb_000245_220 |
0.0885875615 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 14 |
Hb_005000_170 |
0.0886637565 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 15 |
Hb_000075_040 |
0.0892610399 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 16 |
Hb_000749_010 |
0.0894106884 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002400_340 |
0.0897220014 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 18 |
Hb_000254_100 |
0.0897813223 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 19 |
Hb_006483_110 |
0.0900818619 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 20 |
Hb_001153_210 |
0.0912376138 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |