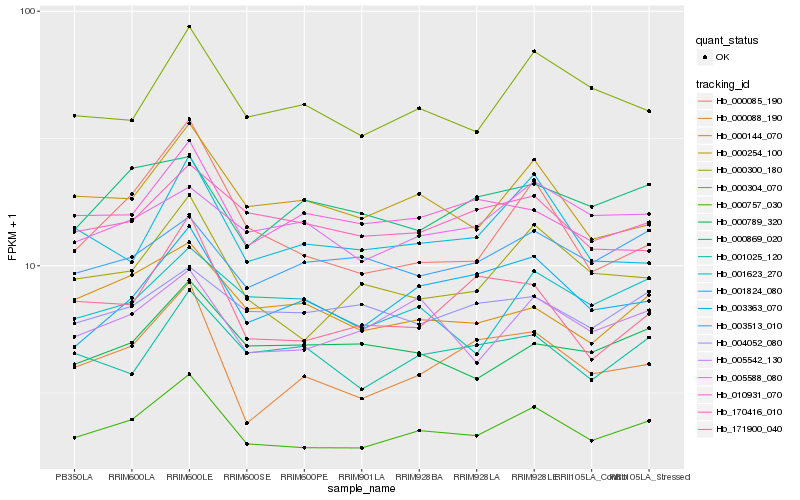
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000757_030 |
0.0 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 2 |
Hb_000300_180 |
0.0690172633 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 3 |
Hb_005542_130 |
0.0725303175 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 4 |
Hb_000254_100 |
0.076750358 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 5 |
Hb_000304_070 |
0.0782570728 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 6 |
Hb_003363_070 |
0.0789278268 |
- |
- |
- |
| 7 |
Hb_010931_070 |
0.0800995926 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000088_190 |
0.0817909548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000144_070 |
0.0841265863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001025_120 |
0.0849348693 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_000789_320 |
0.0861131255 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003513_010 |
0.0874152012 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 13 |
Hb_001623_270 |
0.0877476976 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 14 |
Hb_005588_080 |
0.0908336279 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 15 |
Hb_171900_040 |
0.0912581957 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 16 |
Hb_001824_080 |
0.0916380423 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000869_020 |
0.0923378711 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 18 |
Hb_004052_080 |
0.0923385344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000085_190 |
0.0926757312 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 20 |
Hb_170416_010 |
0.0929705117 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |