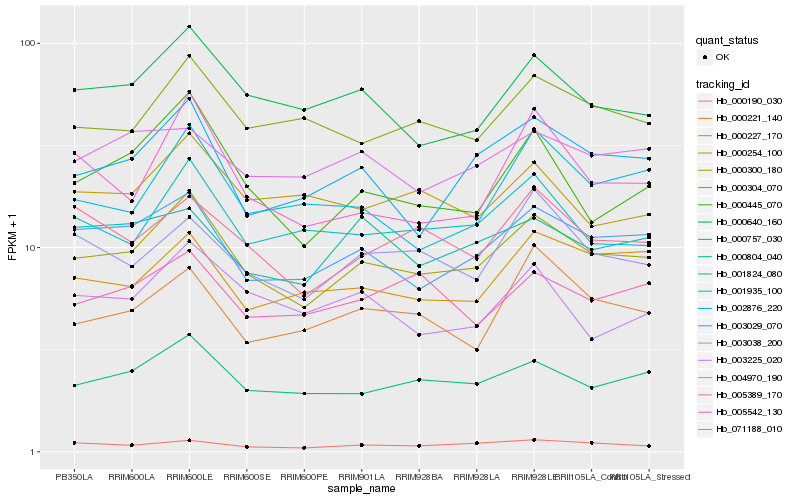
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_180 |
0.0 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 2 |
Hb_000757_030 |
0.0690172633 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 3 |
Hb_003029_070 |
0.06943093 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000640_160 |
0.083313686 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 5 |
Hb_002876_220 |
0.0851480785 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 6 |
Hb_000190_030 |
0.0869389614 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 7 |
Hb_000304_070 |
0.0873329265 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 8 |
Hb_000445_070 |
0.0891080563 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 9 |
Hb_001824_080 |
0.0900735347 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 10 |
Hb_000227_170 |
0.0941181708 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_071188_010 |
0.0941687893 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 12 |
Hb_003038_200 |
0.0967693487 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001935_100 |
0.0996543942 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 14 |
Hb_000804_040 |
0.101263555 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_005542_130 |
0.1015654455 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 16 |
Hb_000254_100 |
0.1025373991 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 17 |
Hb_000221_140 |
0.1027716307 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_005389_170 |
0.1034551502 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 19 |
Hb_004970_190 |
0.1035314974 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_003225_020 |
0.1045120841 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |