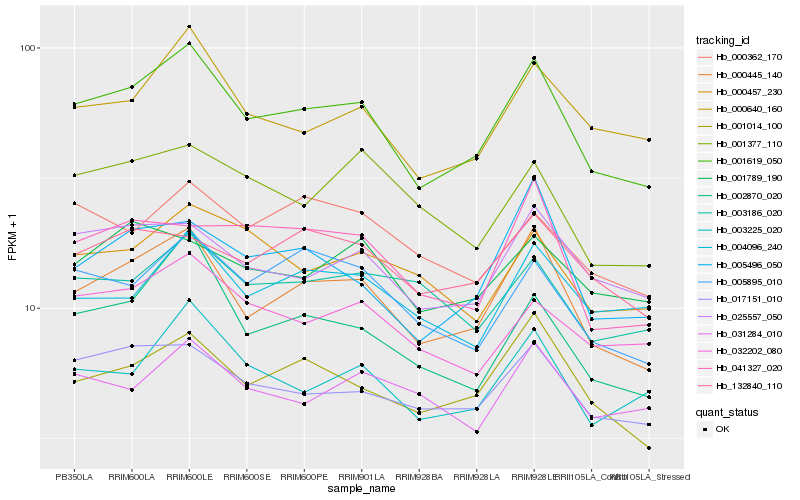
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000445_140 |
0.0399310517 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 3 |
Hb_000640_160 |
0.0720144586 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 4 |
Hb_001014_100 |
0.0722727605 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 5 |
Hb_003225_020 |
0.072844718 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 6 |
Hb_017151_010 |
0.0744289836 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 7 |
Hb_002870_020 |
0.0764840608 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 8 |
Hb_025557_050 |
0.0780297293 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 9 |
Hb_005496_050 |
0.0817391886 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_005895_010 |
0.0820970522 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 11 |
Hb_032202_080 |
0.0844467726 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_041327_020 |
0.0898240796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000457_230 |
0.0900634607 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004096_240 |
0.0911955469 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 15 |
Hb_132840_110 |
0.0916174561 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001377_110 |
0.0930638404 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 17 |
Hb_001789_190 |
0.0937327845 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 18 |
Hb_031284_010 |
0.0940751828 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 19 |
Hb_003186_020 |
0.094226357 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000362_170 |
0.0945285684 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |