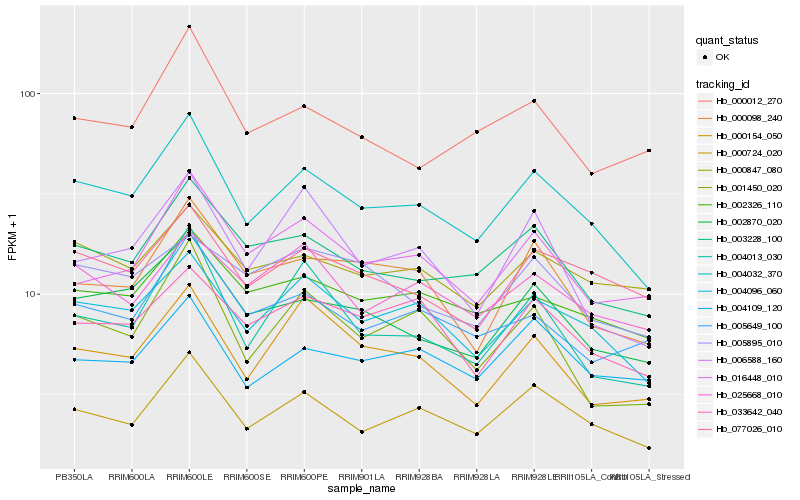
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_370 |
0.0 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 2 |
Hb_004096_060 |
0.0603886472 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000724_020 |
0.0624280947 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 4 |
Hb_002870_020 |
0.0764729995 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003228_100 |
0.0779828926 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 6 |
Hb_002326_110 |
0.0903818955 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 7 |
Hb_077026_010 |
0.0904037646 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 8 |
Hb_000098_240 |
0.0929822445 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 9 |
Hb_004013_030 |
0.0941785378 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 10 |
Hb_033642_040 |
0.0950495777 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001450_020 |
0.095170114 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_004109_120 |
0.0981362742 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005649_100 |
0.0987125357 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000847_080 |
0.0994448748 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_000154_050 |
0.0995361604 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 16 |
Hb_025668_010 |
0.1000630424 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_016448_010 |
0.1008282484 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 18 |
Hb_006588_160 |
0.1063761513 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_000012_270 |
0.1080053837 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 20 |
Hb_005895_010 |
0.1091336177 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |