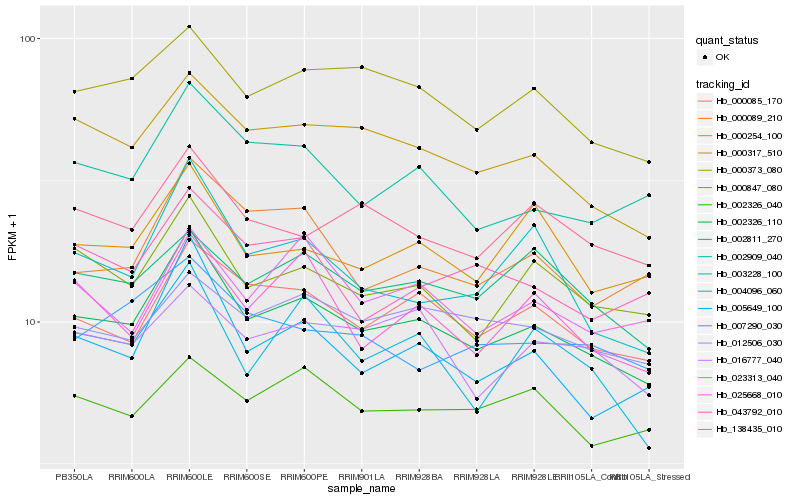
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002326_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 2 |
Hb_005649_100 |
0.0601629343 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000085_170 |
0.0620214441 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000317_510 |
0.0621136175 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 5 |
Hb_000847_080 |
0.0641474223 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_000373_080 |
0.0659912967 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 7 |
Hb_002909_040 |
0.0714053925 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 8 |
Hb_138435_010 |
0.0754632113 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 9 |
Hb_000254_100 |
0.0773758687 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 10 |
Hb_025668_010 |
0.0779978861 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_003228_100 |
0.0784506067 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 12 |
Hb_002811_270 |
0.0797549095 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 13 |
Hb_012506_030 |
0.0801120228 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 14 |
Hb_004096_060 |
0.0805376499 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.08126452 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_016777_040 |
0.081772741 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 17 |
Hb_007290_030 |
0.0830828329 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 18 |
Hb_043792_010 |
0.0831123065 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002326_040 |
0.083220215 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 20 |
Hb_023313_040 |
0.0832803432 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |