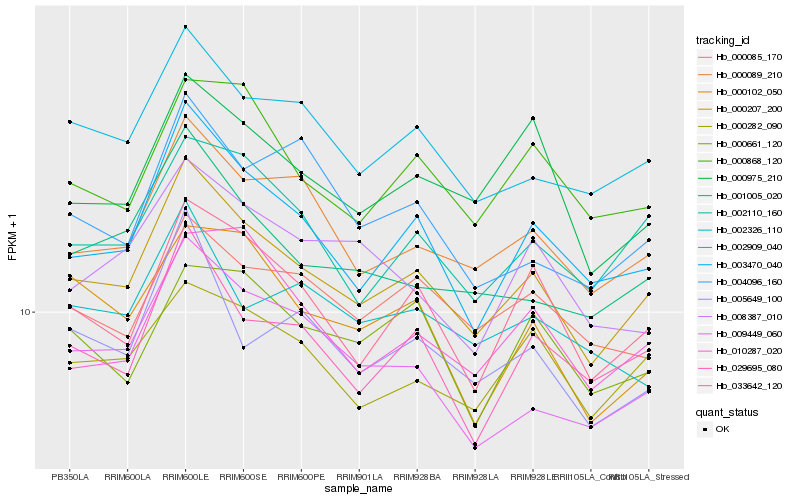
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_200 |
0.0 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_003470_040 |
0.0560166017 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002909_040 |
0.0593799499 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 4 |
Hb_000089_210 |
0.060623336 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_033642_120 |
0.0641819968 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 6 |
Hb_029695_080 |
0.0676013775 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 7 |
Hb_002110_160 |
0.0677538451 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000282_090 |
0.070479032 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 9 |
Hb_001005_020 |
0.0725934945 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 10 |
Hb_000975_210 |
0.0740673924 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 11 |
Hb_000868_120 |
0.0748230787 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 12 |
Hb_000085_170 |
0.0772180228 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005649_100 |
0.0781387608 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 14 |
Hb_009449_060 |
0.0785459111 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 15 |
Hb_010287_020 |
0.0825793435 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 16 |
Hb_004096_160 |
0.0848880074 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 17 |
Hb_000102_050 |
0.0857213384 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002326_110 |
0.0867891827 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 19 |
Hb_000661_120 |
0.0870898521 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 20 |
Hb_008387_010 |
0.0875053662 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |