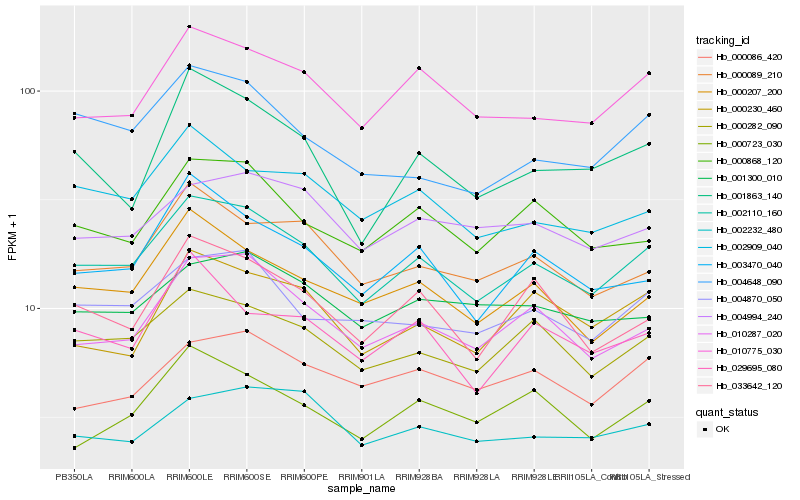
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_160 |
0.0 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000282_090 |
0.0582814944 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 3 |
Hb_010775_030 |
0.0617950424 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 4 |
Hb_010287_020 |
0.0652851239 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 5 |
Hb_000207_200 |
0.0677538451 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_004870_050 |
0.0697311803 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 7 |
Hb_000868_120 |
0.0713680094 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 8 |
Hb_000089_210 |
0.0728367335 |
- |
- |
unknown [Medicago truncatula] |
| 9 |
Hb_033642_120 |
0.0735750122 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 10 |
Hb_002909_040 |
0.0753879176 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 11 |
Hb_003470_040 |
0.0755269378 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_004648_090 |
0.0790462257 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 13 |
Hb_004994_240 |
0.0812791535 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 14 |
Hb_000230_460 |
0.0825134239 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 15 |
Hb_001300_010 |
0.0825404007 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_029695_080 |
0.0854088153 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 17 |
Hb_001863_140 |
0.0854142827 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6b-1 [Jatropha curcas] |
| 18 |
Hb_000086_420 |
0.0854822867 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 19 |
Hb_002232_480 |
0.0855766344 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 20 |
Hb_000723_030 |
0.0860196126 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |