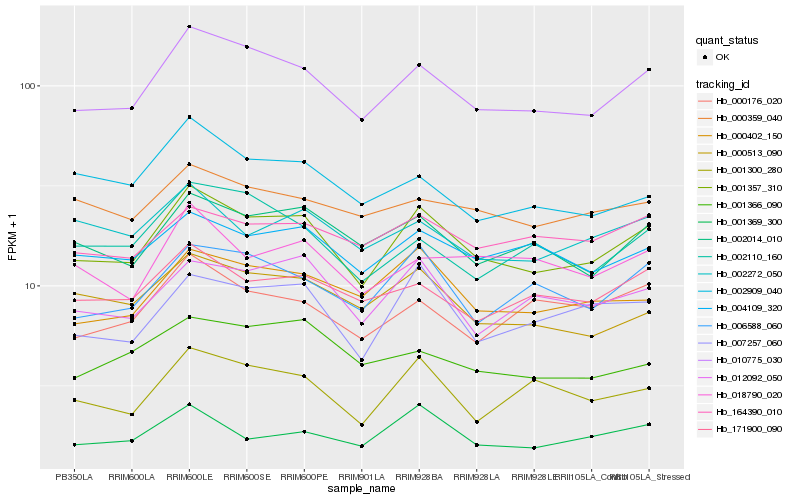
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_310 |
0.0 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_010775_030 |
0.0447889194 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 3 |
Hb_007257_060 |
0.0750925875 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 4 |
Hb_002014_010 |
0.0781049375 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_012092_050 |
0.0784463336 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000402_150 |
0.0790695507 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 7 |
Hb_006588_060 |
0.0811749706 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 8 |
Hb_001369_300 |
0.08282966 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 9 |
Hb_004109_320 |
0.0839854509 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 10 |
Hb_000513_090 |
0.0842942933 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 11 |
Hb_001300_280 |
0.0879032155 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 12 |
Hb_171900_090 |
0.0882107672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002272_050 |
0.0923823909 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 14 |
Hb_164390_010 |
0.0933809964 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 15 |
Hb_018790_020 |
0.0936490119 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002110_160 |
0.093990214 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000176_020 |
0.0956438533 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 18 |
Hb_000359_040 |
0.095737307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002909_040 |
0.0957772456 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 20 |
Hb_001366_090 |
0.0966902964 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |