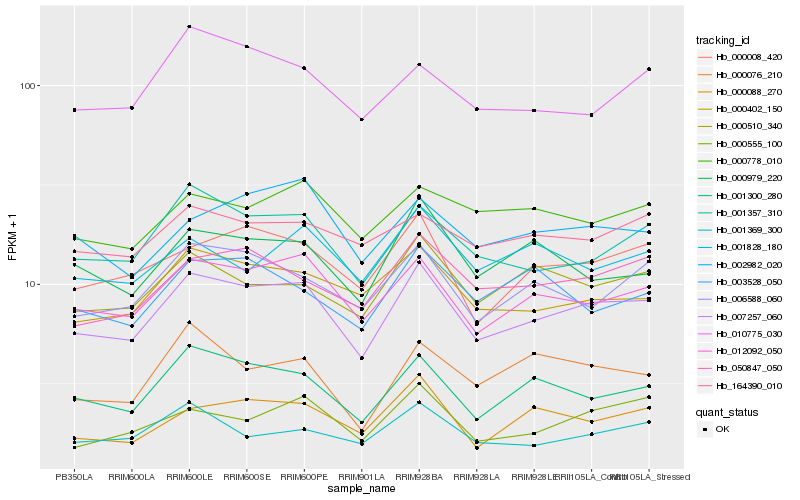
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007257_060 |
0.0 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 2 |
Hb_012092_050 |
0.0590828156 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001300_280 |
0.072017905 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 4 |
Hb_001357_310 |
0.0750925875 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000008_420 |
0.0786404127 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006588_060 |
0.0819300608 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 7 |
Hb_000402_150 |
0.0826179451 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 8 |
Hb_050847_050 |
0.0838913701 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000510_340 |
0.0888596437 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 10 |
Hb_000979_220 |
0.0917147518 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000088_270 |
0.0922700741 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 12 |
Hb_000555_100 |
0.0930025662 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 13 |
Hb_002982_020 |
0.0934976091 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 14 |
Hb_001369_300 |
0.0943475552 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 15 |
Hb_000778_010 |
0.0966228382 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 16 |
Hb_010775_030 |
0.09768418 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 17 |
Hb_000076_210 |
0.0981513143 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001828_180 |
0.0994654959 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 19 |
Hb_003528_050 |
0.0996811522 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_164390_010 |
0.1001340987 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |