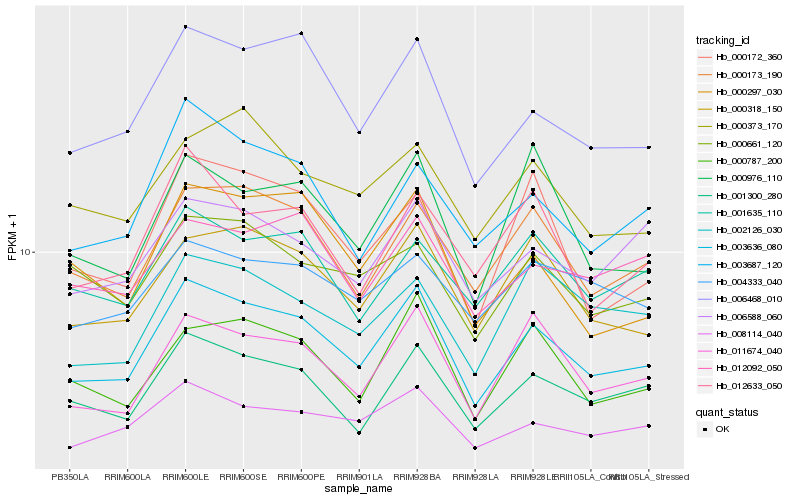
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003636_080 |
0.0 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 2 |
Hb_001300_280 |
0.0673829008 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 3 |
Hb_006468_010 |
0.0680516351 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011674_040 |
0.0692772009 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000173_190 |
0.0697154775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000318_150 |
0.0700747377 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 7 |
Hb_000787_200 |
0.0769352825 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_008114_040 |
0.0836901017 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 9 |
Hb_003687_120 |
0.0840710753 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 10 |
Hb_001635_110 |
0.0873890598 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 11 |
Hb_012633_050 |
0.0879795578 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000976_110 |
0.0888390653 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 13 |
Hb_000172_360 |
0.0894098144 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 14 |
Hb_000373_170 |
0.0899049956 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 15 |
Hb_006588_060 |
0.0901598034 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 16 |
Hb_000297_030 |
0.0907029306 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012092_050 |
0.0915810081 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000661_120 |
0.091613531 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 19 |
Hb_002126_030 |
0.0933040608 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 20 |
Hb_004333_040 |
0.0937131401 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |