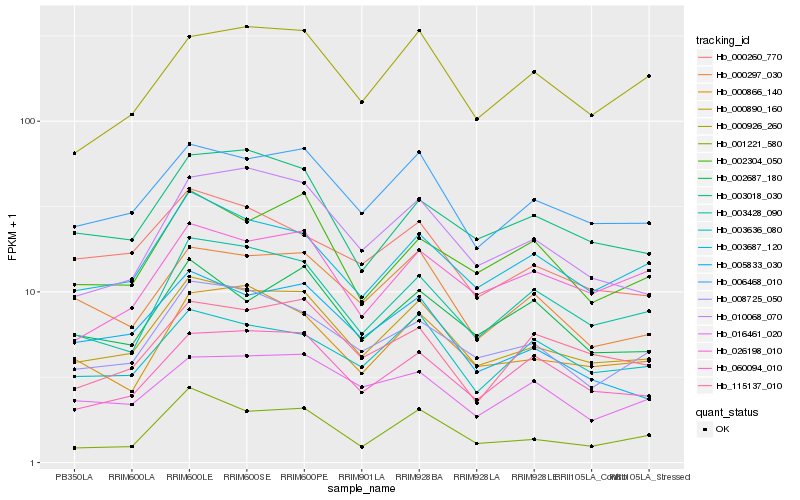
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_160 |
0.0 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 2 |
Hb_006468_010 |
0.0524876225 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008725_050 |
0.0753017991 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_003428_090 |
0.0791519805 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 5 |
Hb_003687_120 |
0.0795019869 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 6 |
Hb_000866_140 |
0.081883875 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 7 |
Hb_010068_070 |
0.0822470993 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000297_030 |
0.0827246165 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003018_030 |
0.0833122733 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 10 |
Hb_060094_010 |
0.0853190902 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000260_770 |
0.0897106328 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 12 |
Hb_002304_050 |
0.0939016958 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 13 |
Hb_016461_020 |
0.0940980896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005833_030 |
0.0944718316 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 15 |
Hb_003636_080 |
0.0950631118 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 16 |
Hb_000926_260 |
0.0962043861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_115137_010 |
0.0973007803 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002687_180 |
0.0977668136 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 19 |
Hb_026198_010 |
0.0988359419 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001221_580 |
0.1007509345 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |