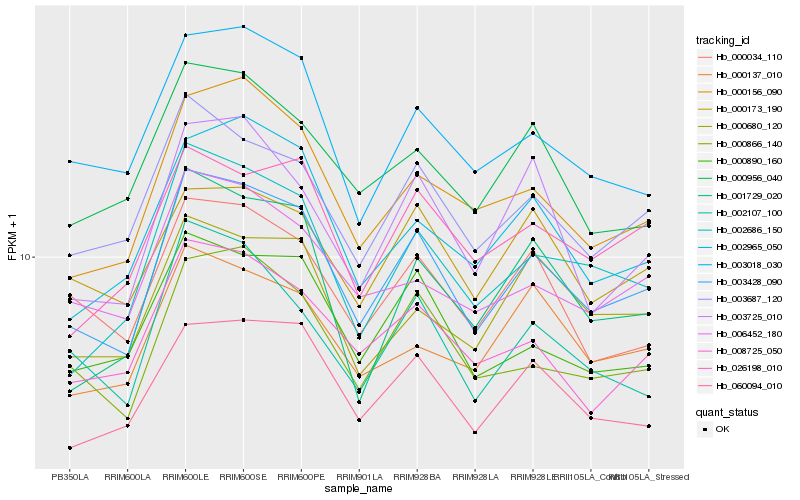
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003428_090 |
0.0 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 2 |
Hb_003687_120 |
0.0717798272 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 3 |
Hb_060094_010 |
0.0719709381 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000866_140 |
0.0732100303 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 5 |
Hb_000156_090 |
0.075608902 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002686_150 |
0.0766428635 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000890_160 |
0.0791519805 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 8 |
Hb_008725_050 |
0.0797954184 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 9 |
Hb_002965_050 |
0.0829358617 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 10 |
Hb_003018_030 |
0.083153568 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 11 |
Hb_000034_110 |
0.0865436159 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 12 |
Hb_026198_010 |
0.0877244439 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000956_040 |
0.0886307513 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 14 |
Hb_002107_100 |
0.0887778454 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_003725_010 |
0.0898104556 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 16 |
Hb_006452_180 |
0.0902334841 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 17 |
Hb_001729_020 |
0.0905401278 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000680_120 |
0.0912016639 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 19 |
Hb_000173_190 |
0.0924526557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000137_010 |
0.0933187184 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |