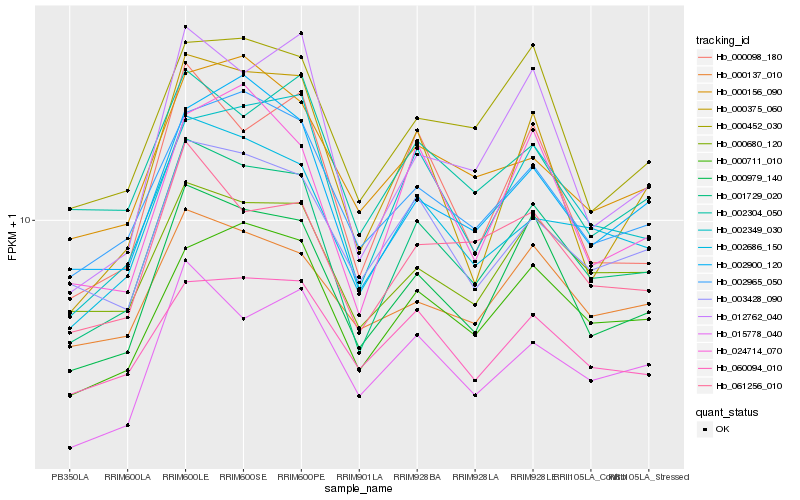
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001729_020 |
0.0 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000979_140 |
0.0705915186 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 3 |
Hb_002686_150 |
0.0734803328 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_000711_010 |
0.08664878 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 5 |
Hb_002349_030 |
0.0881875678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003428_090 |
0.0905401278 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 7 |
Hb_002965_050 |
0.09249667 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 8 |
Hb_000680_120 |
0.0949263015 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 9 |
Hb_061256_010 |
0.0964767334 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002900_120 |
0.0989835165 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 11 |
Hb_000098_180 |
0.1019940514 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 12 |
Hb_060094_010 |
0.1031889601 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 13 |
Hb_015778_040 |
0.1032979013 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 14 |
Hb_000375_060 |
0.1041524886 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 15 |
Hb_000156_090 |
0.1061810894 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_012762_040 |
0.1063942718 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 17 |
Hb_000137_010 |
0.1066000438 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 18 |
Hb_024714_070 |
0.1070424517 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 19 |
Hb_000452_030 |
0.1075372485 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_002304_050 |
0.1078423837 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |