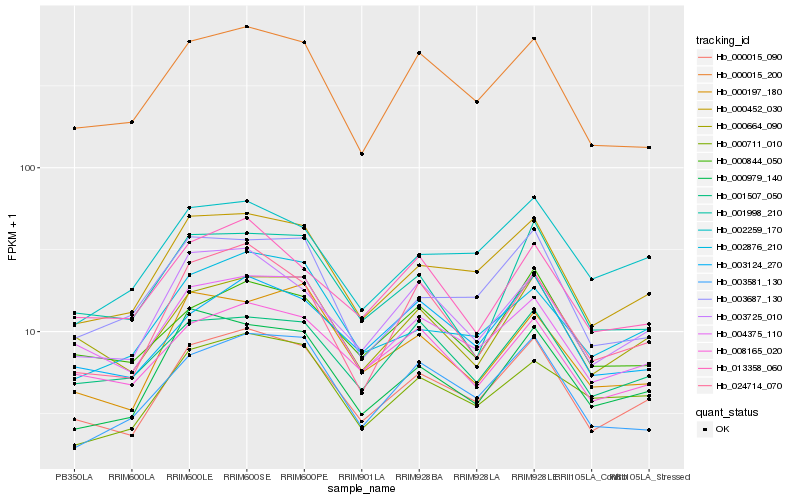
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_090 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 2 |
Hb_000452_030 |
0.057788163 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_000664_090 |
0.0776556371 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 4 |
Hb_004375_110 |
0.0804493457 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 5 |
Hb_003725_010 |
0.0824579811 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 6 |
Hb_000015_200 |
0.0852944176 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 7 |
Hb_000979_140 |
0.087094502 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 8 |
Hb_024714_070 |
0.08835232 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 9 |
Hb_002876_210 |
0.0934972455 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 10 |
Hb_003581_130 |
0.0946061667 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_008165_020 |
0.094683991 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003124_270 |
0.0954051958 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 13 |
Hb_000711_010 |
0.0974140541 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 14 |
Hb_003687_130 |
0.0990868238 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 15 |
Hb_002259_170 |
0.1009472702 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001507_050 |
0.1011772846 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000844_050 |
0.1012516448 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 18 |
Hb_001998_210 |
0.1031066254 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000197_180 |
0.103133614 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 20 |
Hb_013358_060 |
0.1031753103 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |