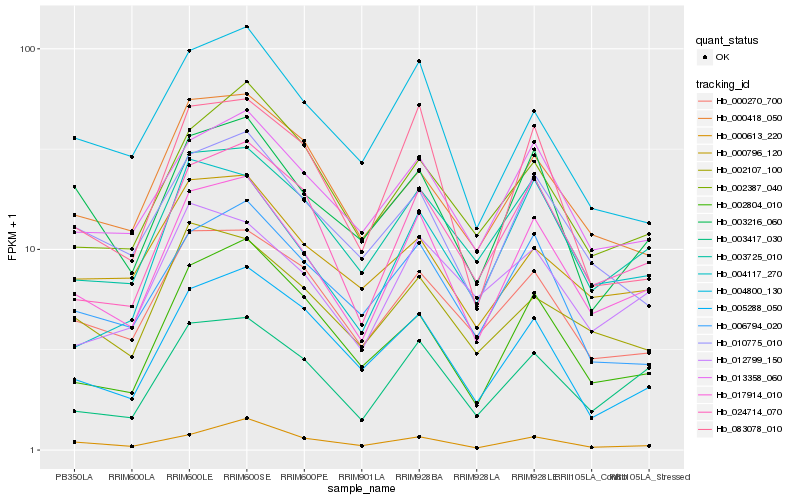
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017914_010 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 2 |
Hb_024714_070 |
0.0798661305 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 3 |
Hb_003417_030 |
0.0814295038 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003725_010 |
0.0854869151 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 5 |
Hb_013358_060 |
0.0909845217 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 6 |
Hb_000270_700 |
0.1020854612 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 7 |
Hb_010775_010 |
0.1063858496 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003216_060 |
0.1074600327 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 9 |
Hb_006794_020 |
0.1120082931 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 10 |
Hb_002804_010 |
0.1127089594 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 11 |
Hb_083078_010 |
0.112880835 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 12 |
Hb_002107_100 |
0.1131538059 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_004117_270 |
0.1132775073 |
- |
- |
- |
| 14 |
Hb_002387_040 |
0.1137732684 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_012799_150 |
0.1141300034 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methyl-CpG-binding domain-containing protein 13 [Jatropha curcas] |
| 16 |
Hb_000418_050 |
0.115462298 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 17 |
Hb_005288_050 |
0.1172479188 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 18 |
Hb_000796_120 |
0.1181189963 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000613_220 |
0.1200771217 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_004800_130 |
0.120790016 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |