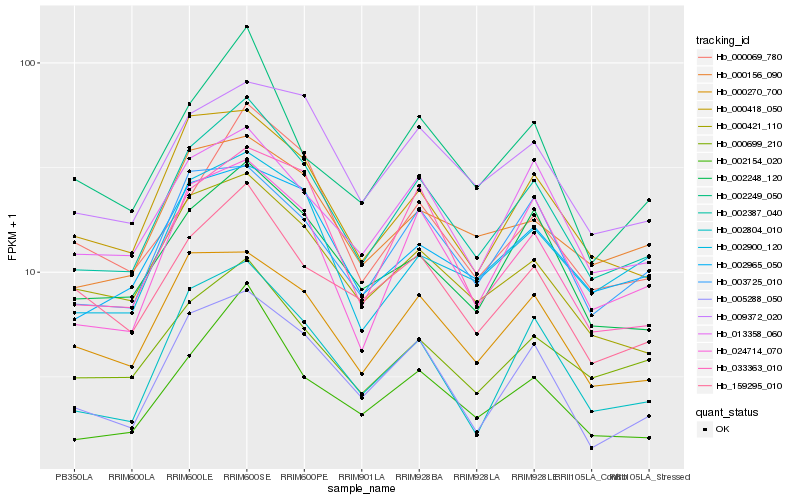
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002387_040 |
0.0 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 2 |
Hb_002248_120 |
0.0812860255 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 3 |
Hb_000699_210 |
0.0824705143 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 4 |
Hb_024714_070 |
0.0869924338 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 5 |
Hb_000418_050 |
0.0893150271 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 6 |
Hb_005288_050 |
0.0898467163 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 7 |
Hb_002804_010 |
0.0903150809 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 8 |
Hb_013358_060 |
0.0903313048 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 9 |
Hb_000421_110 |
0.0905973038 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002965_050 |
0.0920068012 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 11 |
Hb_003725_010 |
0.092390737 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 12 |
Hb_002154_020 |
0.0932359223 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_009372_020 |
0.093236947 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 14 |
Hb_000270_700 |
0.0932618082 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 15 |
Hb_002900_120 |
0.0945598961 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 16 |
Hb_033363_010 |
0.0960047655 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 17 |
Hb_159295_010 |
0.0963069097 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 18 |
Hb_000156_090 |
0.0971591693 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000069_780 |
0.1000489688 |
- |
- |
atpob1, putative [Ricinus communis] |
| 20 |
Hb_002249_050 |
0.1002925387 |
- |
- |
Auxin response factor, putative [Ricinus communis] |