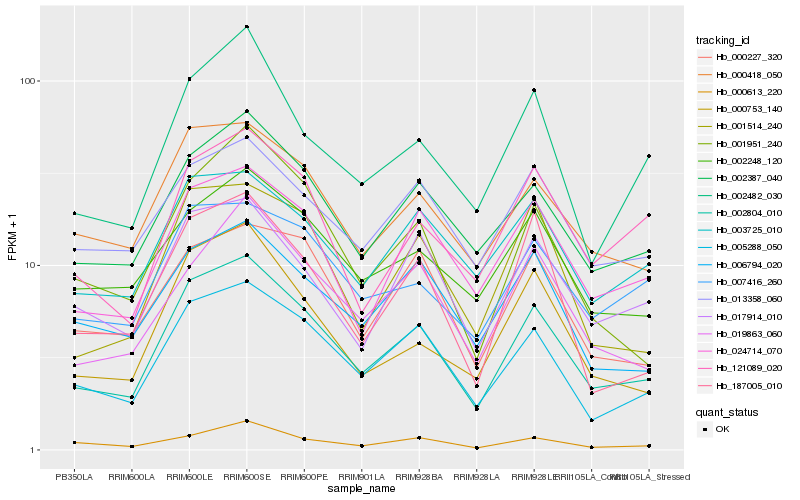
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002804_010 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 2 |
Hb_005288_050 |
0.0763691442 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 3 |
Hb_002387_040 |
0.0903150809 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 4 |
Hb_000418_050 |
0.0985852388 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 5 |
Hb_024714_070 |
0.1019918391 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 6 |
Hb_000753_140 |
0.1067189819 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 7 |
Hb_001951_240 |
0.1069399949 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 8 |
Hb_001514_240 |
0.1074522643 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_019863_060 |
0.1084015908 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 10 |
Hb_000613_220 |
0.1091000882 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 11 |
Hb_002482_030 |
0.110784283 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 12 |
Hb_017914_010 |
0.1127089594 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 13 |
Hb_002248_120 |
0.1128333794 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_187005_010 |
0.1158142836 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 15 |
Hb_007416_260 |
0.1166390852 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 16 |
Hb_003725_010 |
0.1184054911 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 17 |
Hb_006794_020 |
0.1189373919 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 18 |
Hb_000227_320 |
0.1205549577 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_121089_020 |
0.1208479283 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 20 |
Hb_013358_060 |
0.1215678425 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |