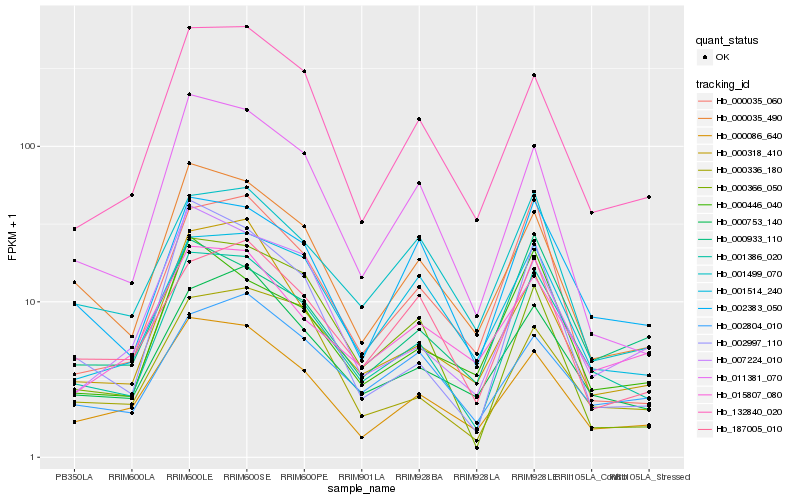
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001386_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000086_640 |
0.1072680256 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_000753_140 |
0.108733638 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 4 |
Hb_132840_020 |
0.1151822511 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 5 |
Hb_000035_490 |
0.118680357 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 6 |
Hb_002383_050 |
0.1222370085 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 7 |
Hb_011381_070 |
0.1291759565 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 8 |
Hb_001514_240 |
0.1303562687 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_000318_410 |
0.1314674506 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_001499_070 |
0.1315333468 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 11 |
Hb_000336_180 |
0.1321209641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_187005_010 |
0.1322331827 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 13 |
Hb_000035_060 |
0.1330492651 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000446_040 |
0.1360259377 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 15 |
Hb_007224_010 |
0.1366171906 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 16 |
Hb_002804_010 |
0.1374808572 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 17 |
Hb_015807_080 |
0.1403120511 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 18 |
Hb_000933_110 |
0.1414271994 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 19 |
Hb_000366_050 |
0.1435020219 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_002997_110 |
0.1446995253 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |