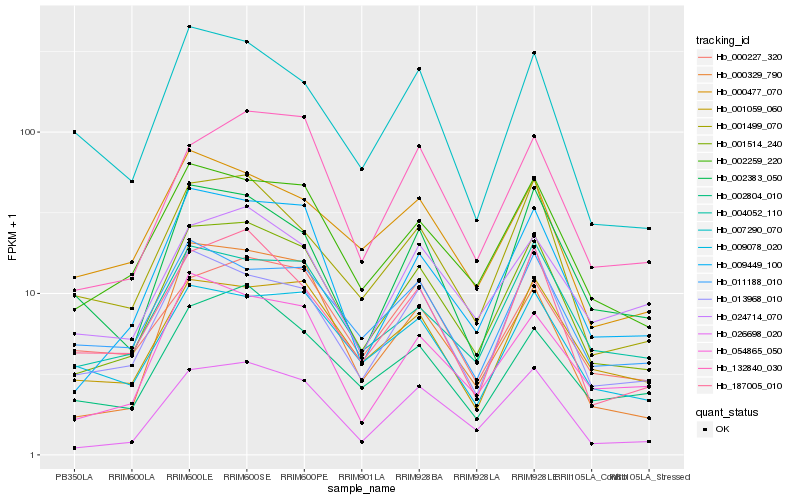
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_240 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 2 |
Hb_026698_020 |
0.0724306651 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_002259_220 |
0.0750378349 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 4 |
Hb_009449_100 |
0.0819590797 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 5 |
Hb_001499_070 |
0.0921892017 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 6 |
Hb_013968_010 |
0.0990880188 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 7 |
Hb_187005_010 |
0.1008334989 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 8 |
Hb_004052_110 |
0.1008410465 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_132840_030 |
0.10238029 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 10 |
Hb_001059_060 |
0.1038145142 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 11 |
Hb_054865_050 |
0.1064166404 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002804_010 |
0.1074522643 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 13 |
Hb_000329_790 |
0.1120809893 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 14 |
Hb_009078_020 |
0.1126421392 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 15 |
Hb_002383_050 |
0.1137440706 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 16 |
Hb_024714_070 |
0.1155734529 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 17 |
Hb_011188_010 |
0.1158252937 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000477_070 |
0.1163612038 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 19 |
Hb_000227_320 |
0.1174713493 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_007290_070 |
0.1176586175 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |