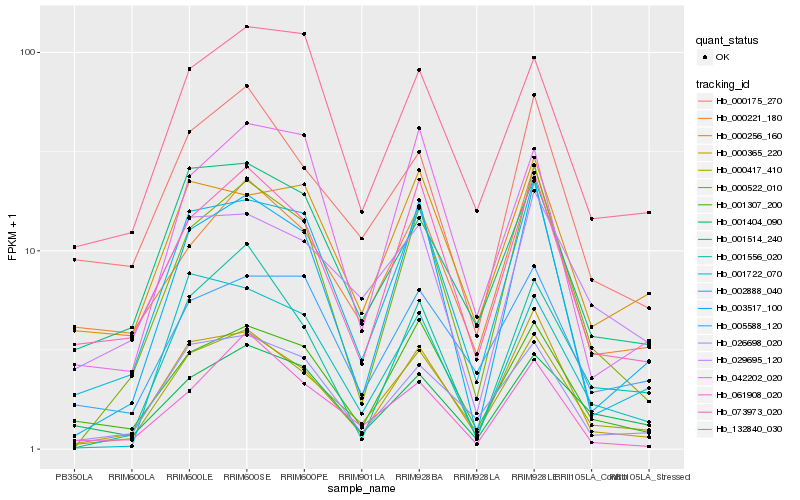
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_410 |
0.0 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000365_220 |
0.0915744597 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 3 |
Hb_026698_020 |
0.1006096027 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 4 |
Hb_073973_020 |
0.1047382623 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 5 |
Hb_002888_040 |
0.1142814021 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 6 |
Hb_001307_200 |
0.1145597993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 7 |
Hb_000522_010 |
0.1181211036 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 8 |
Hb_001514_240 |
0.1252854892 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_003517_100 |
0.1275822043 |
transcription factor |
TF Family: GRAS |
- |
| 10 |
Hb_000175_270 |
0.1308016845 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001556_020 |
0.1349656196 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 12 |
Hb_001722_070 |
0.1358961268 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 13 |
Hb_005588_120 |
0.1381162949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001404_090 |
0.1389009783 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 15 |
Hb_000221_180 |
0.1395349949 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 16 |
Hb_132840_030 |
0.1440072042 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 17 |
Hb_029695_120 |
0.1449348927 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 18 |
Hb_000256_160 |
0.1471970622 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 19 |
Hb_061908_020 |
0.1473054705 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 20 |
Hb_042202_020 |
0.1473554282 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |