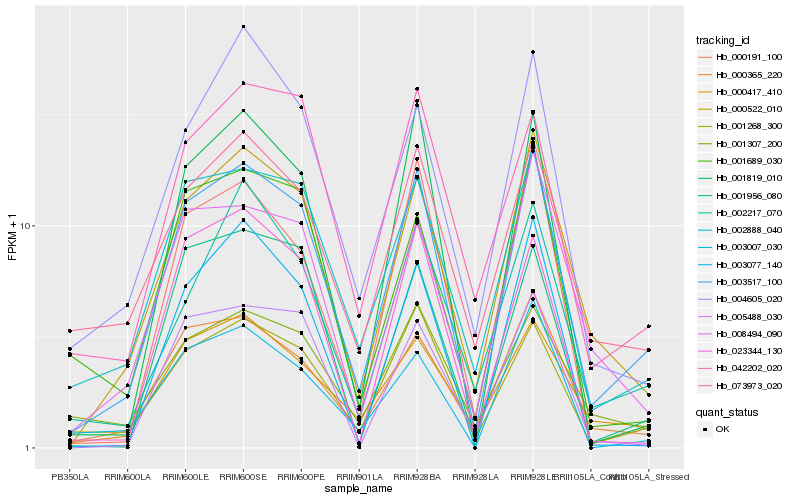
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003517_100 |
0.0 |
transcription factor |
TF Family: GRAS |
- |
| 2 |
Hb_001268_300 |
0.1015505509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000522_010 |
0.1034148662 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 4 |
Hb_002888_040 |
0.1060683082 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 5 |
Hb_000365_220 |
0.1070828905 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 6 |
Hb_000417_410 |
0.1275822043 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_008494_090 |
0.131730627 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 8 |
Hb_001819_010 |
0.1335324206 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 9 |
Hb_000191_100 |
0.1367104886 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_073973_020 |
0.1387617639 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 11 |
Hb_003007_030 |
0.140012869 |
- |
- |
- |
| 12 |
Hb_042202_020 |
0.1412501278 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 13 |
Hb_001689_030 |
0.1417419762 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 14 |
Hb_023344_130 |
0.1430831889 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_001956_080 |
0.1437172557 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 16 |
Hb_002217_070 |
0.144825587 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 17 |
Hb_005488_030 |
0.1450564353 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 18 |
Hb_001307_200 |
0.1461480356 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 19 |
Hb_004605_020 |
0.1473380306 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 20 |
Hb_003077_140 |
0.148649547 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |