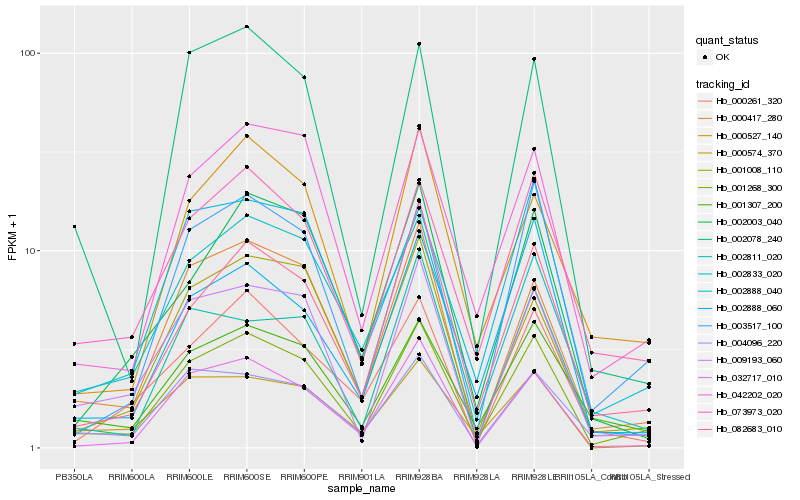
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_300 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003517_100 |
0.1015505509 |
transcription factor |
TF Family: GRAS |
- |
| 3 |
Hb_000417_280 |
0.1032831071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_042202_020 |
0.105789388 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 5 |
Hb_009193_060 |
0.1065393428 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 6 |
Hb_082683_010 |
0.1075489164 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_073973_020 |
0.1075717349 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 8 |
Hb_001307_200 |
0.109501465 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 9 |
Hb_002888_040 |
0.1123167102 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 10 |
Hb_004096_220 |
0.1139654815 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_032717_010 |
0.1169860597 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001008_110 |
0.1218041509 |
- |
- |
- |
| 13 |
Hb_002833_020 |
0.1236827994 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_000527_140 |
0.1285221857 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_000574_370 |
0.1332918545 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_020 |
0.1333354027 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 17 |
Hb_002078_240 |
0.1334232396 |
rubber biosynthesis |
Gene Name: SRPP4 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 18 |
Hb_002003_040 |
0.1370539297 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002888_060 |
0.137542101 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000261_320 |
0.1381515196 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |