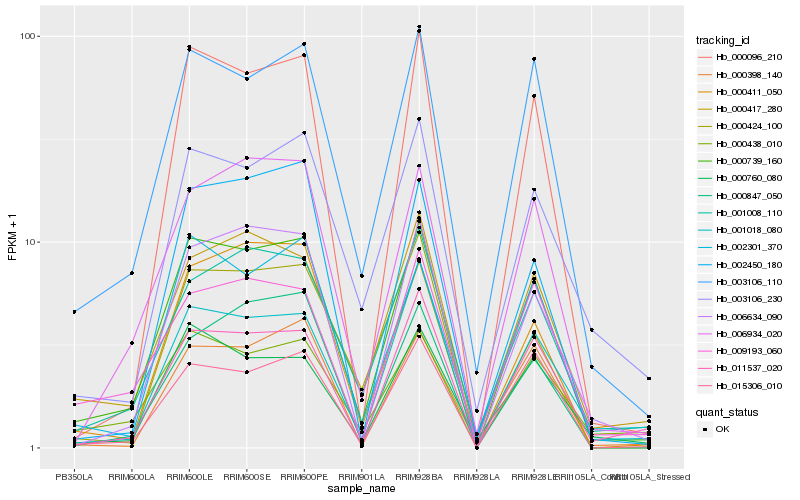
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_160 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_003106_110 |
0.0869610662 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000096_210 |
0.0996067216 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 4 |
Hb_006634_090 |
0.1042593066 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 5 |
Hb_006934_020 |
0.1071086821 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 6 |
Hb_002301_370 |
0.1074365118 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 7 |
Hb_000417_280 |
0.1105659249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001008_110 |
0.1145718941 |
- |
- |
- |
| 9 |
Hb_011537_020 |
0.1146783468 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 10 |
Hb_015306_010 |
0.116096295 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 11 |
Hb_000411_050 |
0.1196154749 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 12 |
Hb_000438_010 |
0.1224348684 |
- |
- |
- |
| 13 |
Hb_001018_080 |
0.122771383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000760_080 |
0.1228655435 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 15 |
Hb_003106_230 |
0.1249425437 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 16 |
Hb_000398_140 |
0.1261233763 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 17 |
Hb_000424_100 |
0.1267836125 |
desease resistance |
Gene Name: G-alpha |
GTP-binding protein (p) alpha subunit, gpa1, putative [Ricinus communis] |
| 18 |
Hb_002450_180 |
0.1286672637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000847_050 |
0.128859268 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 20 |
Hb_009193_060 |
0.1288831898 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |