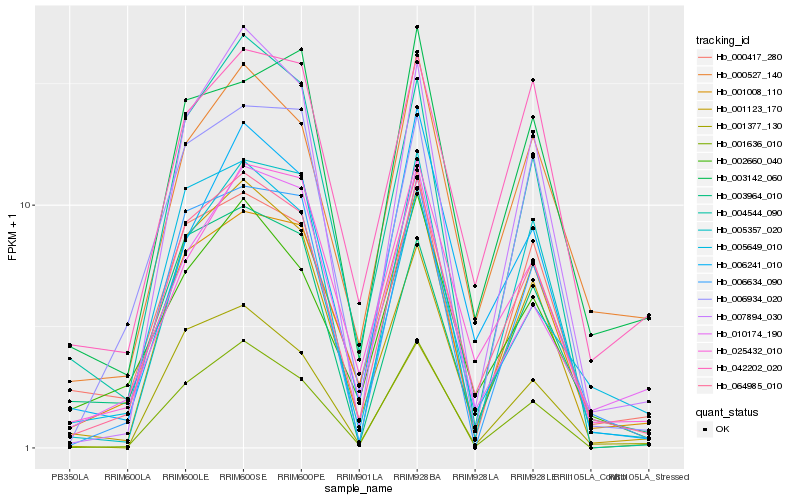
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064985_010 |
0.0 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 2 |
Hb_001008_110 |
0.0983650036 |
- |
- |
- |
| 3 |
Hb_007894_030 |
0.1075895364 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_006634_090 |
0.1085292726 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 5 |
Hb_001636_010 |
0.1094141226 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 6 |
Hb_000527_140 |
0.1095821327 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 7 |
Hb_005649_010 |
0.1137257106 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 8 |
Hb_025432_010 |
0.1139580748 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 9 |
Hb_003964_010 |
0.1142279868 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 10 |
Hb_002660_040 |
0.1150499509 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001123_170 |
0.1189215933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000417_280 |
0.1193357363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006934_020 |
0.1196127835 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 14 |
Hb_004544_090 |
0.1220515989 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_010174_190 |
0.1222781622 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 16 |
Hb_042202_020 |
0.1246850738 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 17 |
Hb_006241_010 |
0.1250432772 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_005357_020 |
0.1258197353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003142_060 |
0.1287027654 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001377_130 |
0.1287075897 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |