| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
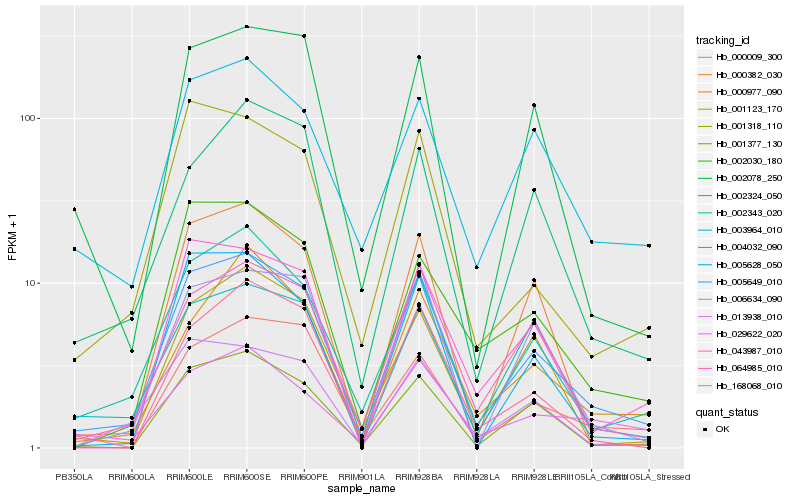
Hb_005649_010 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 2 |
Hb_002343_020 |
0.1088076153 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 3 |
Hb_003964_010 |
0.1117554009 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 4 |
Hb_064985_010 |
0.1137257106 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 5 |
Hb_001377_130 |
0.1159361185 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 6 |
Hb_013938_010 |
0.1261621478 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 7 |
Hb_001318_110 |
0.1308053885 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 8 |
Hb_005628_050 |
0.1330706391 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_000382_030 |
0.1344218252 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 10 |
Hb_002078_250 |
0.1354382908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004032_090 |
0.1364545021 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 12 |
Hb_006634_090 |
0.1372217855 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 13 |
Hb_168068_010 |
0.137712859 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 14 |
Hb_043987_010 |
0.1385342671 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 15 |
Hb_001123_170 |
0.1389260251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000009_300 |
0.1395535259 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 17 |
Hb_002030_180 |
0.1400902482 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_002324_050 |
0.1401069081 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 19 |
Hb_000977_090 |
0.1401574613 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 20 |
Hb_029622_020 |
0.1407932341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |