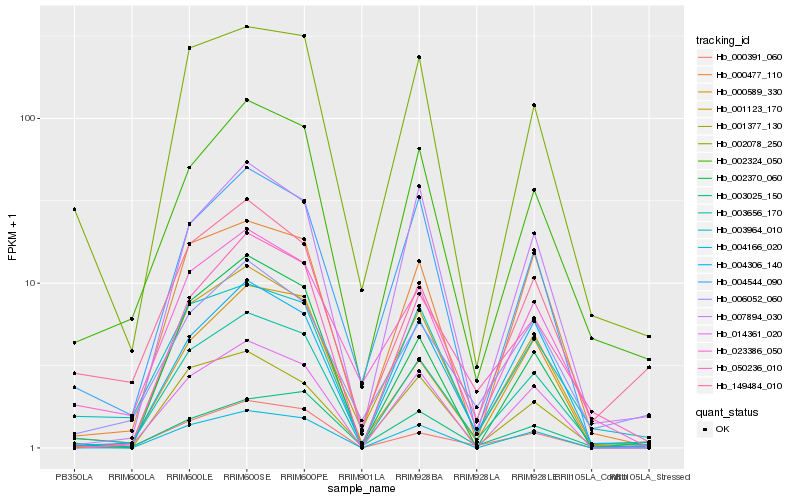
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003656_170 |
0.0 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 2 |
Hb_001123_170 |
0.0745303057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002324_050 |
0.0821640553 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 4 |
Hb_002078_250 |
0.0991381077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003964_010 |
0.1016373961 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 6 |
Hb_004306_140 |
0.1091147417 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_050236_010 |
0.1130642952 |
- |
- |
hypothetical protein EUGRSUZ_E02960, partial [Eucalyptus grandis] |
| 8 |
Hb_000391_060 |
0.1146246514 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 9 |
Hb_000477_110 |
0.1155275709 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 10 |
Hb_003025_150 |
0.1167029192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004544_090 |
0.1196945075 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_023386_050 |
0.119830486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001377_130 |
0.1211182461 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 14 |
Hb_014361_020 |
0.1225536418 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 15 |
Hb_000589_330 |
0.1230533386 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_006052_060 |
0.1236231178 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_002370_060 |
0.124286701 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 18 |
Hb_149484_010 |
0.1246592019 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 19 |
Hb_004166_020 |
0.1253141494 |
- |
- |
- |
| 20 |
Hb_007894_030 |
0.1253326537 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |