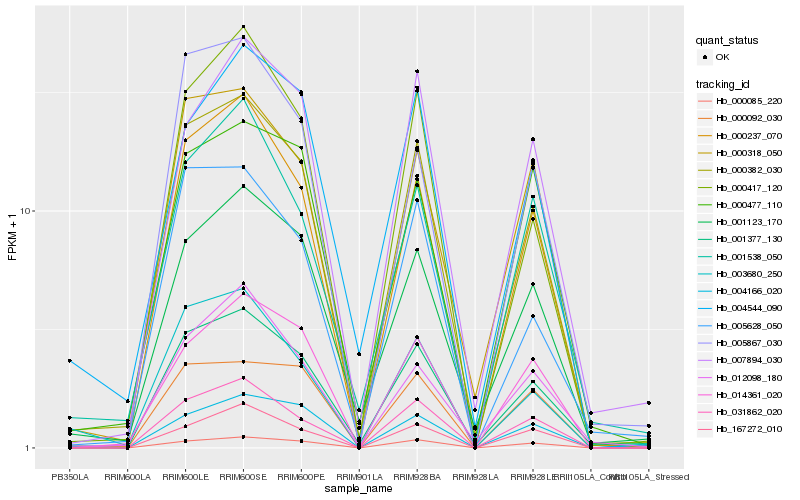
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000382_030 |
0.0 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 2 |
Hb_001377_130 |
0.0676811788 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 3 |
Hb_000237_070 |
0.0757866246 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 4 |
Hb_000318_050 |
0.0880407144 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000085_220 |
0.092419948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_031862_020 |
0.0928933726 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 7 |
Hb_001123_170 |
0.0939290427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000417_120 |
0.0995264533 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 9 |
Hb_005628_050 |
0.105325699 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_014361_020 |
0.1062079992 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 11 |
Hb_001538_050 |
0.1139652664 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 12 |
Hb_000092_030 |
0.1159640016 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 13 |
Hb_004166_020 |
0.1159723812 |
- |
- |
- |
| 14 |
Hb_167272_010 |
0.1167800143 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 15 |
Hb_007894_030 |
0.118503793 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_003680_250 |
0.1190009579 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 17 |
Hb_005867_030 |
0.1197035642 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 18 |
Hb_004544_090 |
0.1204686483 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000477_110 |
0.1206194848 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 20 |
Hb_012098_180 |
0.121058487 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |