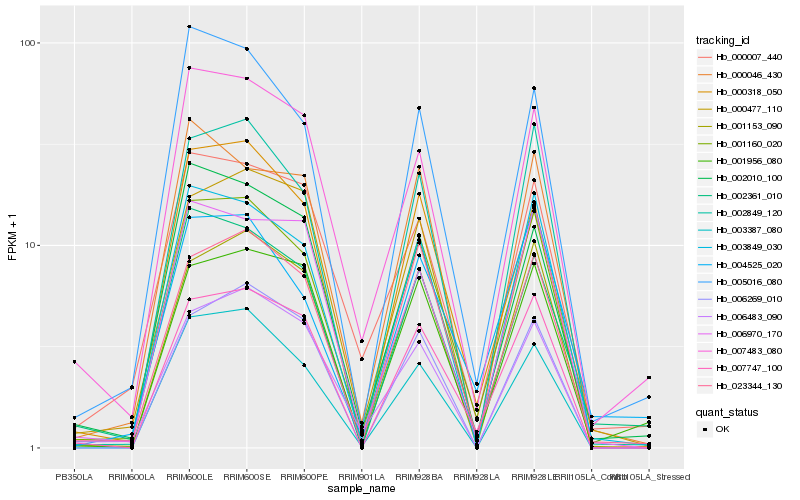
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_020 |
0.0 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 2 |
Hb_007747_100 |
0.0759191688 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 3 |
Hb_003849_030 |
0.0776277679 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 4 |
Hb_002849_120 |
0.0857932634 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_001153_090 |
0.0881792844 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 6 |
Hb_000318_050 |
0.0891969206 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 7 |
Hb_023344_130 |
0.0977735297 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_003387_080 |
0.0982327548 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_005016_080 |
0.1068921862 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 10 |
Hb_006483_090 |
0.1137902221 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 11 |
Hb_000046_430 |
0.1146306942 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 12 |
Hb_007483_080 |
0.1165172653 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 13 |
Hb_006970_170 |
0.1176405055 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_006269_010 |
0.1184227233 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 15 |
Hb_002010_100 |
0.1190691672 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 16 |
Hb_002361_010 |
0.1198315524 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000477_110 |
0.121503436 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 18 |
Hb_001956_080 |
0.1219370078 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 19 |
Hb_000007_440 |
0.124369158 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 20 |
Hb_004525_020 |
0.1247149538 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |