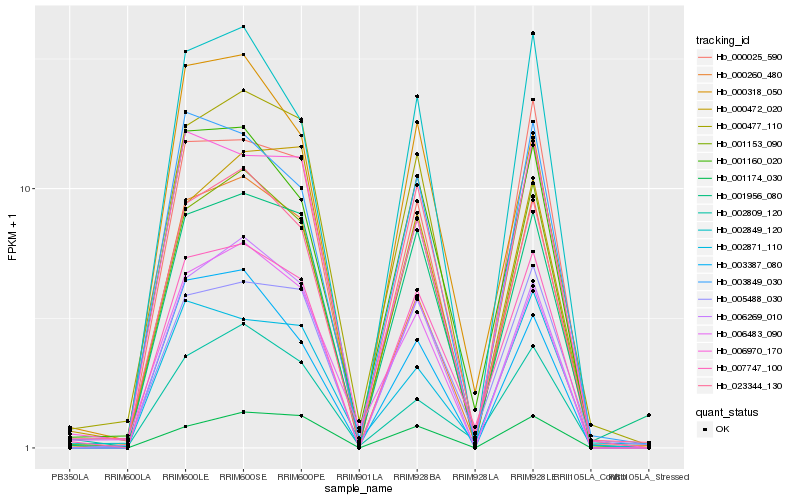
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_100 |
0.0 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 2 |
Hb_001160_020 |
0.0759191688 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 3 |
Hb_001153_090 |
0.0821638986 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 4 |
Hb_005488_030 |
0.0957755433 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 5 |
Hb_006269_010 |
0.1011089553 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 6 |
Hb_002849_120 |
0.1019119795 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 7 |
Hb_001956_080 |
0.1024503912 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 8 |
Hb_006970_170 |
0.105474444 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_003849_030 |
0.1075715811 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 10 |
Hb_023344_130 |
0.1098860536 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000318_050 |
0.1130383968 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000260_480 |
0.1136303884 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 13 |
Hb_000477_110 |
0.1140397249 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 14 |
Hb_000025_590 |
0.1157956722 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001174_030 |
0.1202733944 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002871_110 |
0.1255111008 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 17 |
Hb_002809_120 |
0.1257996506 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 18 |
Hb_006483_090 |
0.1273802676 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_000472_020 |
0.1275386162 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 20 |
Hb_003387_080 |
0.1290774831 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |