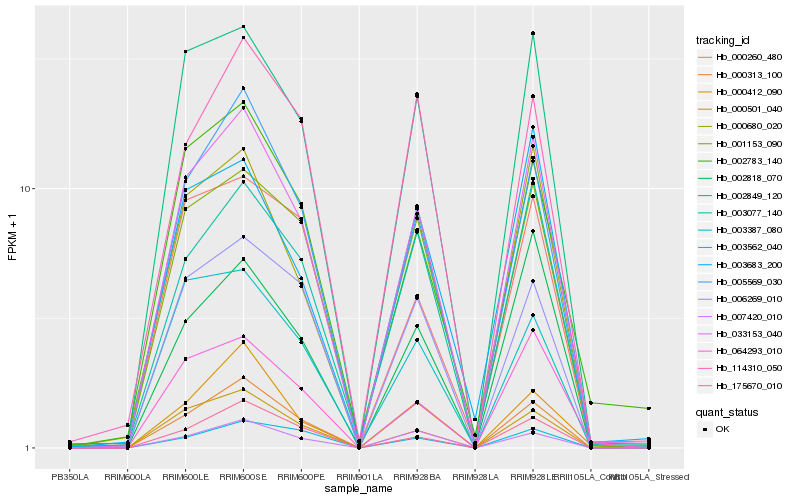
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033153_040 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002849_120 |
0.0692826327 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 3 |
Hb_005569_030 |
0.0711193023 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000680_020 |
0.0888251131 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_003077_140 |
0.0894949098 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 6 |
Hb_000313_100 |
0.0946103073 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 7 |
Hb_001153_090 |
0.0956256061 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 8 |
Hb_064293_010 |
0.1023149345 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_175670_010 |
0.1088486982 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_006269_010 |
0.1105818871 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 11 |
Hb_002818_070 |
0.1138735688 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_002783_140 |
0.1146697538 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 13 |
Hb_007420_010 |
0.1186515413 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 14 |
Hb_114310_050 |
0.121329786 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 15 |
Hb_003387_080 |
0.1214786037 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000501_040 |
0.1253100404 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000412_090 |
0.1255471494 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 18 |
Hb_003562_040 |
0.1260678722 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_003683_200 |
0.1270319981 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 20 |
Hb_000260_480 |
0.1300620974 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |