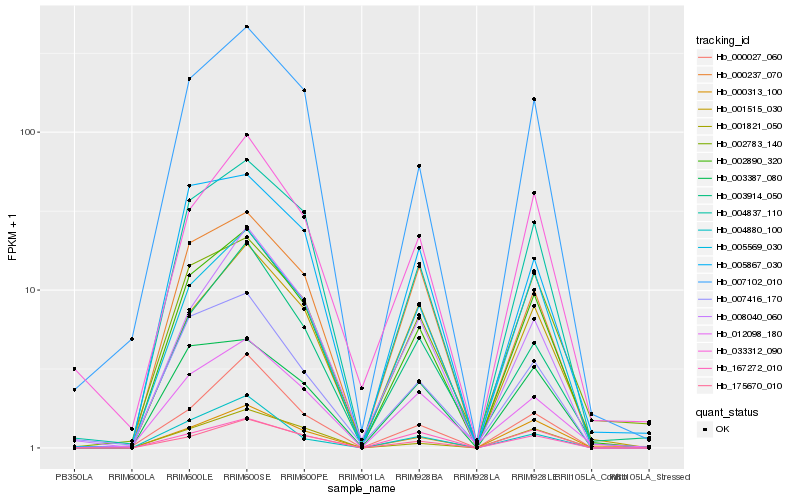
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 2 |
Hb_004837_110 |
0.0657093002 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007102_010 |
0.0839323359 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 4 |
Hb_005569_030 |
0.0892787192 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000313_100 |
0.0993156306 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 6 |
Hb_012098_180 |
0.1089721903 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 7 |
Hb_000237_070 |
0.1136158815 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 8 |
Hb_002783_140 |
0.1143806601 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 9 |
Hb_001821_050 |
0.1149902323 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 10 |
Hb_001515_030 |
0.1157119917 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_175670_010 |
0.1172827503 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_007416_170 |
0.1175752193 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 13 |
Hb_003914_050 |
0.1185009147 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 14 |
Hb_000027_060 |
0.1210112267 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 15 |
Hb_003387_080 |
0.1210588435 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_008040_060 |
0.1211681448 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 17 |
Hb_033312_090 |
0.1218862874 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005867_030 |
0.1247204655 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 19 |
Hb_004880_100 |
0.1270655065 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 20 |
Hb_167272_010 |
0.1279051609 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |