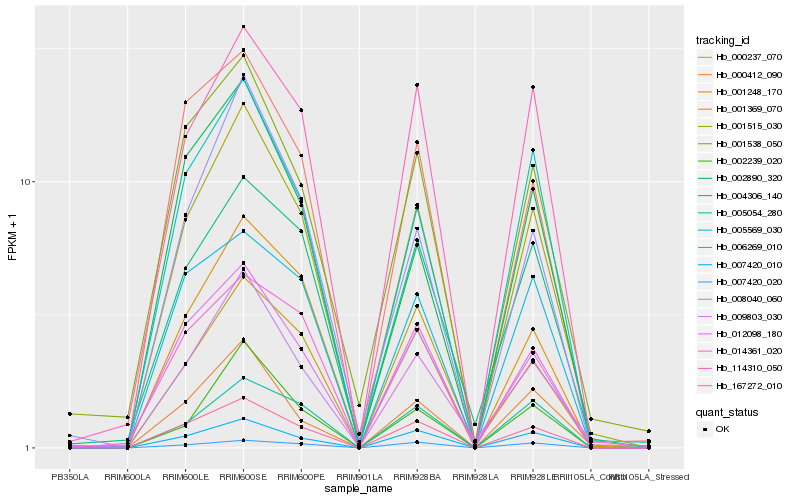
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001515_030 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_167272_010 |
0.0626205885 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 3 |
Hb_009803_030 |
0.087982067 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 4 |
Hb_007420_010 |
0.0882854437 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005569_030 |
0.0914762826 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 6 |
Hb_114310_050 |
0.0941349463 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 7 |
Hb_008040_060 |
0.102017168 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_000412_090 |
0.1052690963 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 9 |
Hb_001248_170 |
0.1062122369 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 10 |
Hb_005054_280 |
0.1096938149 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 11 |
Hb_014361_020 |
0.1133672679 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 12 |
Hb_012098_180 |
0.1141346627 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 13 |
Hb_004306_140 |
0.114173649 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_000237_070 |
0.1147858743 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 15 |
Hb_002890_320 |
0.1157119917 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 16 |
Hb_001538_050 |
0.1160162689 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 17 |
Hb_006269_010 |
0.1167637527 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 18 |
Hb_002239_020 |
0.1189201657 |
- |
- |
MLO, putative [Theobroma cacao] |
| 19 |
Hb_001369_070 |
0.1190091515 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_007420_020 |
0.1200401979 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |