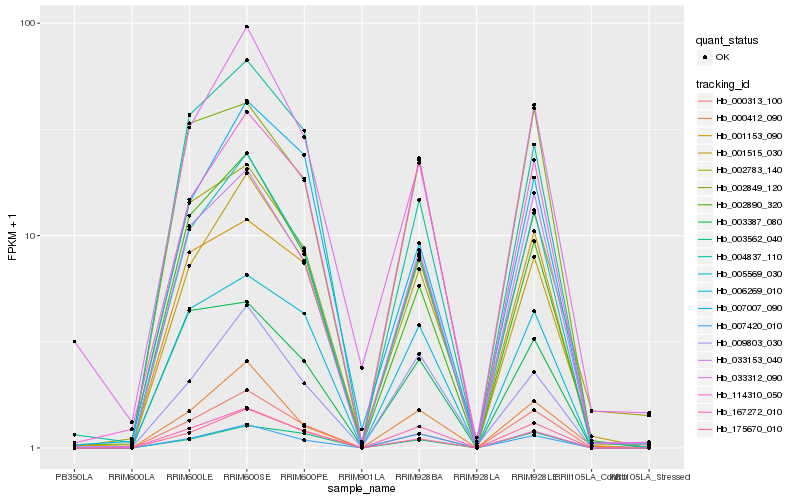
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005569_030 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000313_100 |
0.0650992806 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 3 |
Hb_033153_040 |
0.0711193023 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_175670_010 |
0.0774857874 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_002890_320 |
0.0892787192 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 6 |
Hb_001515_030 |
0.0914762826 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_167272_010 |
0.1016724392 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 8 |
Hb_006269_010 |
0.1055618359 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 9 |
Hb_004837_110 |
0.1062914026 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000412_090 |
0.1077549076 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_033312_090 |
0.1079776267 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_007420_010 |
0.1086207496 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003562_040 |
0.1119651133 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_002783_140 |
0.1150666912 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 15 |
Hb_114310_050 |
0.116784055 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 16 |
Hb_002849_120 |
0.1188772 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 17 |
Hb_001153_090 |
0.1214386969 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 18 |
Hb_003387_080 |
0.1232407278 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_007007_090 |
0.1267152529 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 20 |
Hb_009803_030 |
0.1272743132 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |