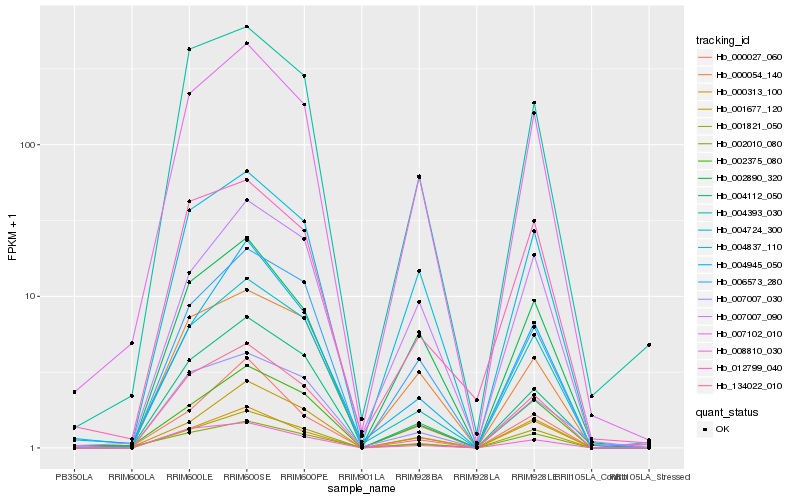
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007102_010 |
0.0 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 2 |
Hb_004837_110 |
0.0615833644 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001821_050 |
0.0768179987 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 4 |
Hb_002890_320 |
0.0839323359 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 5 |
Hb_002375_080 |
0.0884552595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004724_300 |
0.0907215317 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 7 |
Hb_004112_050 |
0.0944057952 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_004393_030 |
0.095469667 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 9 |
Hb_001677_120 |
0.0985250262 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_002010_080 |
0.1036368887 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 11 |
Hb_006573_280 |
0.1072093226 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 12 |
Hb_007007_090 |
0.1103628862 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 13 |
Hb_134022_010 |
0.1141587113 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 14 |
Hb_000054_140 |
0.11460192 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 15 |
Hb_008810_030 |
0.1153014839 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_012799_040 |
0.1187195849 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 17 |
Hb_000027_060 |
0.1217198573 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 18 |
Hb_000313_100 |
0.122412927 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 19 |
Hb_007007_030 |
0.1254030448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004945_050 |
0.1259072156 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |