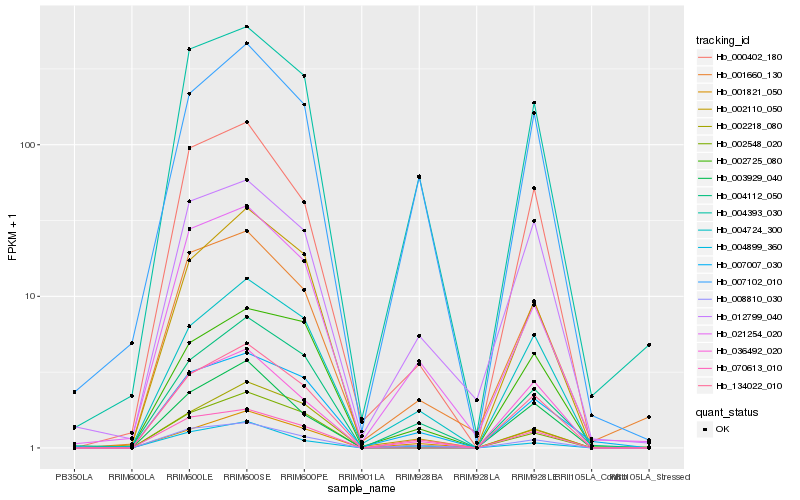
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134022_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 2 |
Hb_004724_300 |
0.0802940157 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 3 |
Hb_008810_030 |
0.0825921327 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000402_180 |
0.0872620486 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 5 |
Hb_036492_020 |
0.0903145575 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_001821_050 |
0.091804594 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 7 |
Hb_004393_030 |
0.0967389989 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 8 |
Hb_002110_050 |
0.097442291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004112_050 |
0.1014157301 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_002218_080 |
0.1031027001 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 11 |
Hb_070613_010 |
0.1035663669 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_001660_130 |
0.1090161557 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 13 |
Hb_007102_010 |
0.1141587113 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 14 |
Hb_021254_020 |
0.1148047117 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_007007_030 |
0.1179163784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012799_040 |
0.1181227522 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 17 |
Hb_003929_040 |
0.1186830027 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004899_360 |
0.1206482864 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_002548_020 |
0.1230524779 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002725_080 |
0.123281602 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |