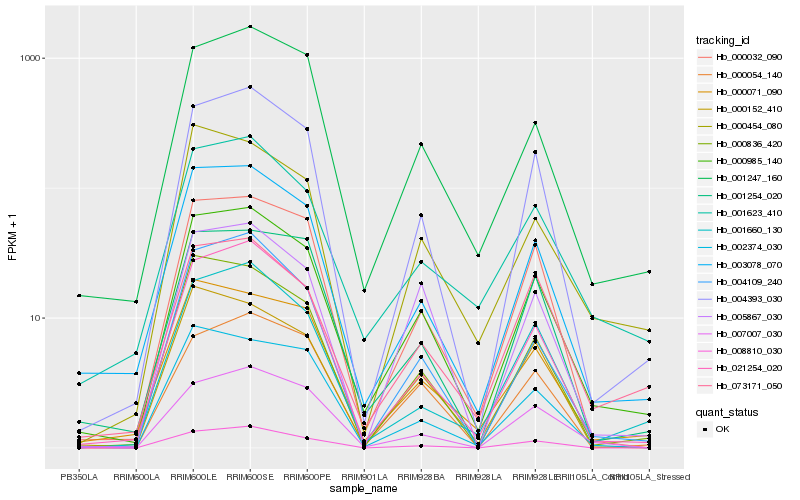
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000985_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 2 |
Hb_004393_030 |
0.0823760768 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 3 |
Hb_003078_070 |
0.0831718863 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 4 |
Hb_021254_020 |
0.0881225115 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 5 |
Hb_001247_160 |
0.0950675211 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 6 |
Hb_000032_090 |
0.0988578365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000071_090 |
0.1015310992 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 8 |
Hb_000836_420 |
0.1032079512 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001623_410 |
0.1075173538 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004109_240 |
0.1096682173 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_007007_030 |
0.1100402514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008810_030 |
0.1101374045 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001660_130 |
0.1146396433 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 14 |
Hb_000454_080 |
0.1159104701 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 15 |
Hb_000152_410 |
0.1173106028 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_001254_020 |
0.117439809 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_073171_050 |
0.1176087019 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 18 |
Hb_005867_030 |
0.1183130723 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 19 |
Hb_000054_140 |
0.1215206363 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 20 |
Hb_002374_030 |
0.1216393012 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |