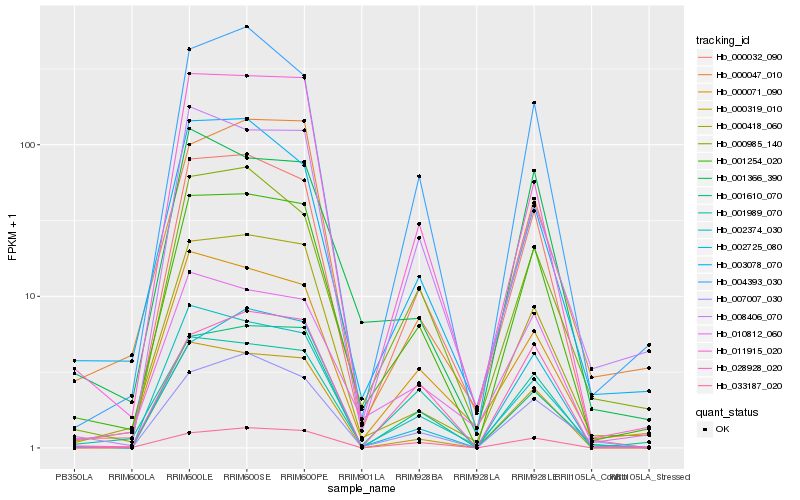
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001254_020 |
0.0 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_000032_090 |
0.0783134722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000418_060 |
0.0982063354 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 4 |
Hb_004393_030 |
0.1092400844 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 5 |
Hb_007007_030 |
0.1099976109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000071_090 |
0.1109959011 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 7 |
Hb_001610_070 |
0.1118953554 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 8 |
Hb_002374_030 |
0.1119435685 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000319_010 |
0.1124091778 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 10 |
Hb_010812_060 |
0.1145085793 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 11 |
Hb_011915_020 |
0.1173106116 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000985_140 |
0.117439809 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 13 |
Hb_003078_070 |
0.1187672006 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 14 |
Hb_008406_070 |
0.1218273097 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 15 |
Hb_001989_070 |
0.1239779067 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 16 |
Hb_033187_020 |
0.1241606687 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 17 |
Hb_002725_080 |
0.1248895308 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_000047_010 |
0.1251891149 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 19 |
Hb_028928_020 |
0.1252774745 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 20 |
Hb_001366_390 |
0.1254714227 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |