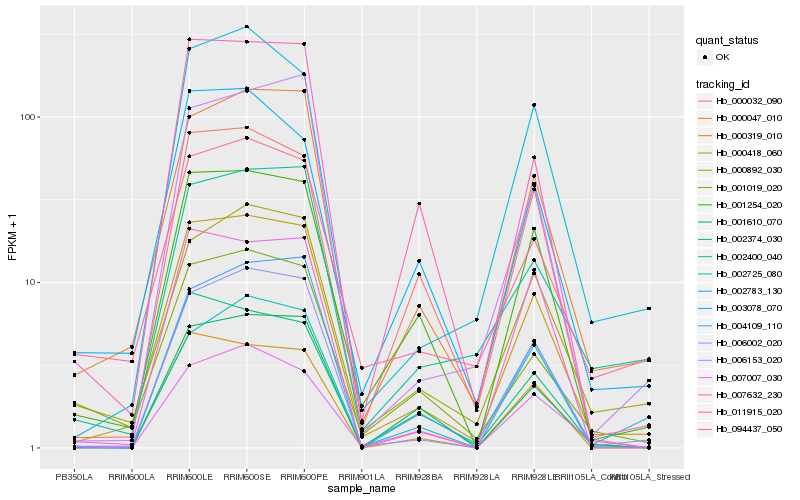
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_060 |
0.0 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 2 |
Hb_006002_020 |
0.0776378876 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 3 |
Hb_000047_010 |
0.0776858803 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 4 |
Hb_001254_020 |
0.0982063354 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 5 |
Hb_000319_010 |
0.1090562381 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 6 |
Hb_011915_020 |
0.1092984698 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002783_130 |
0.1123139897 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000892_030 |
0.1164745937 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 9 |
Hb_006153_020 |
0.1186872197 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 10 |
Hb_094437_050 |
0.1191336994 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002374_030 |
0.1204783285 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000032_090 |
0.1207504544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004109_110 |
0.1228499428 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 14 |
Hb_001019_020 |
0.124807865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001610_070 |
0.124866939 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 16 |
Hb_007632_230 |
0.126181657 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 17 |
Hb_002725_080 |
0.1265850913 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_003078_070 |
0.1275078432 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 19 |
Hb_002400_040 |
0.1284850027 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 20 |
Hb_007007_030 |
0.1295931098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |